Part IV: Understanding and Navigating Y SNP Results
This page may be embedded in an HTML frame. If so, be sure to utilize the vertical scroll bar on the HTML frame as well as the vertical scroll bar in your browser. On the right side of the browser window, the frame scrollbar should be left of your browser scrollbar.
Prerequisites: Parts I and and III are recommended.
- Is there a relationship or correlation between STR markers and SNPs?
- What is FTDNA's yDNA Haplotree?
- My known first/second/third paternal line cousins has done SNP testing and we match on STRs. Do I need to test SNPs too?
- My terminal SNP appears in an SNP stream that aligns with the SNP stream of others in my surname project, but mine remains stuck upstream from the terminal SNPs of other members. Why?
- How are SNPs named?
- Why should I get SNP testing done?
- What are the major parent Y haplogroups?
- Can different parent Y haplogroups experience the same SNP mutation?
- What kinds of Y SNP testing are available at FTDNA?
- How do I access my Big Y results?
- How are my Big Y results integrated into the Y Haplotree?
- How do I look at ONLY my Y STR matches who have tested Big Y?
- Is it possible that a SNP match and I could share a terminal SNP and we are *not* STR matches, even though we share a surname and geographic paternal origins?
- Is it possible for matches sharing my terminal SNP branch to be missing upstream mutations?
- Does FTDNA publish date formation or TMRCA estimates of SNPs?
- What is a no-call?
- What can I do with my SNP results?
- What is FTDNA's Big Y Matches Tool?
- What is FTDNA's Block Tree Tool?
- I had Big Y 500 done on a relative but the test is no longer offered. Will FTDNA still support the results?
- How does FTDNA build the Y haplotree?
-
The short answer is SOMETIMES. There may be some correlations because both the SNP mutations and the STR mutations occurred so long ago that they look like they occurred about the same time. You may recall that in Part III, there was a discussion about the relationship between haplotype and haplogroup with a discussion of correlation between the two.
Correlation is not causation. SNP mutations and STR mutations occur independently of each other. Looking at SNP and STR mutations that occurred about the same time is somewhat like looking at two stars in the sky next to each other, but in reality are many millions of miles apart.
Although SNP and STR mutations occur independently of one another, some haplogroups can be predicted from STRs because of their appearance from a distance. Likewise there are plenty of haplogroups that cannot be predicted from STR, especially in R. With that as a preface you would need to discuss diffusion and extinction as processes.
Here is a silly analogy. Consider a pond and somebody pours a gallon of ink in the center. That ink in time spreads out by diffusion, the center is the modal and radius from center is genetic distance. STR mutations can be represented by ink concentration and further off center the lower is the concentration. Until some other process kicks in everybody at a given radius has the same STR.
Now imagine a bird flies over and lets loose with poop that all lands in one spot at random in the ever growing ink circle. The poop represents a one time event such as a SNP mutation and it is measured not by concentration but is it in a water sample or not. Right after the poop first drops, other water samples equidistant from the center have the same concentration of ink but no poop. So in this case you cannot predict the existence of poop by measuring ink concentration, analogous to to being unable to predict a haplogroup from STR. As time continues the water soluble poop also spreads by diffusion.
Now some time later the fickle finger of fate enters the picture in the form of extinction. A man with a special vacuum cleaner sucks out the infected water nearly everywhere but carelessly leaves behind two blobs; one poop infected sampled from one radius and another not infected from a significantly different radius. Finally along comes a kid with his science kit and he discovers that poop free water has 100 times less ink it than contaminated water. Hence he has found a way to predict that water is contaminated without having to measure the poop (SNP); one simply has to measure the ink (STR). The kid wins a blue ribbon at his school's science fair.
Haplogroups that are young can't be predicted from off modal STR, the SNP has to be tested, like right after the bird pooped you can't predict the existence of poop from the ink, you have to have a poop test as well. In the old haplogroups there has been enough time for extinction to kick in and now the SNP of subgroups can be predicted from the STR.
-
The FTDNA Y Haplotree is FTDNA's representation of the Y DNA haplotree. It is populated from FTDNA's SNP database which is produced from customer Big Y Advanced SNP test results.
In your account dashboard, under Partner Applications and Other Tools, there is a widget called Public Haplotrees. You can access both the Y and Mitochondrial Haplotrees from that widget.
-
No. If you have close Y surname matches at a high number of markers (111) AND you know how you are related by conventional genealogy methods (i.e., paper record trail), you can consider yourself done with Y DNA testing at least in terms of recent genetic genealogy research if your interest does not extend further.
If one of you has done SNP testing, you can probably safely conclude that you are in that same SNP branch and you don't all need to deep test SNPs. One of you can serve as an SNP proxy for the other cousins.
-
The image on the right shows real data from a surname project. Project members with terminal SNPs I-A13665/I-Y13631 and I-A13664 have done advanced SNP testing, but have not seen their terminal SNPs change since testing. Most of the surname project members in this haplogroup fall downstream of those testers, somewhere under I-A14359.
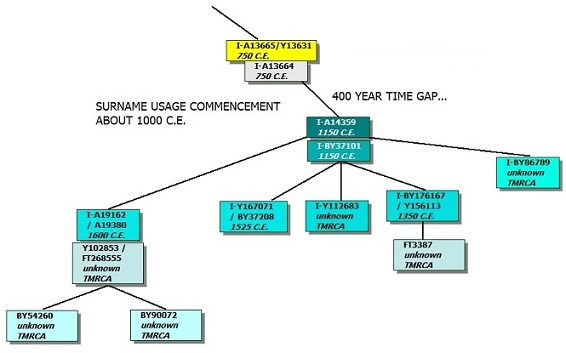
By formation date estimates (at the time this tutorial was written), I-A13665 came into being about 300 C.E., with a TMRCA about 850 C.E. I-A13664 may have been formed around 850 C.E. This is over 1200 years ago. These testers did their DNA tests in the 21st century, not a millennium ago. What gives?
These lineages did not stop spawning new SNPs a millennium ago. They kept right on developing new mutations. We have not identified those mutations yet. Those testers have private variants that may not be recognized yet as other branches.
REASON A. Failure to identify newer mutations could be due to the lack of other data (sufficient numbers of testers) to compare to. We might see a cousin branch to I-A14359 develop.
REASON B. Our knowledge of this branch of the haplotree is imperfect. With more data in the future, the Y haplotree as we see it now might be recognized as erroneous; statistics would get recalculated and these SNPs could get repositioned, or even dropped altogether with others to replace them.
REASON C. There could also have been a DNA copy problem from a father to son that has been retained in successive generations. If testers show a good fit in a subclade of A14359 but lack the A14359 mutation itself, that could be where the copy error occurred.
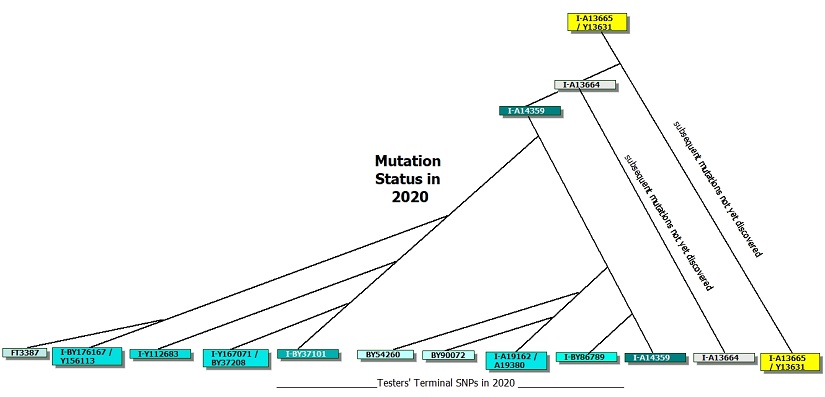
The diagram below is a different view of the mutation history diagram above. It is a status of mutation discoveries. The significance of private mutation variants among these testers is not yet understood.
-
SNP names get long forms and short forms.
ISOGG is the organization that defines the long forms, which are patterned to show the relationships of SNPs to each other. Long form names are expected to change over time as new information is learned.
Think of the long form as a street address. Occasionally a city grows to a point at which it needs to renumber its streets, and some of the residents get new addresses as a result.
In the same way, a haplotree can grow and expand as new subclades are discovered. New statistical information may reveal a slightly altered pedigree for an SNP. As a consequence the long forms get revised. So don't get too fond of a long form because it could be temporary.
Below are examples of long form SNPs with their short form names, stacked up so you can see their relationship to each other:
R1b1a1b1a1a2c1 R-L21
R1b1a1b1a1a2c1a R-DF13
R1b1a1b1a1a2c1a3 R-FGC11134
R1b1a1b1a1a2c1a3a R-Z16250
R1b1a1b1a1a2c1a3a2 R-CTS4466Notice how R-DF13, a child of R-L21, includes the long form of R-L21 in its own long form. As you work down the list, you'll see how the long form of the next child is built on the long form of its parent SNP.
The name in a short form starts with a capital letter, followed by a dash, followed by more letters and then a number. The letter preceding the dash is the parent haplogroup, such as I or E or J or R.
The initial letters after the dash in a short form help to identify the lab that discovered that mutation. For example, an SNP short form starting with "Y" was discovered by the YFull lab in Russia. An SNP short form starting with "BY" was discovered at the FTDNA lab through the Big Y test. ISOGG has pages on SNP naming you might find interesting.
When the context is understood, you might see the initial capital letter and the dash omitted, for example I-A13665 could be just A13665 or R-CTS4466 is just CTS4466.
SNPs might also have multiple short form variants. Several labs could observe the same mutation at the same Y chromosome location, and then give the mutation its own name. Over time, those variants are recognized.
Short form names are permanent. Their names do not carry the structural relationship of SNPs to each other.
-
It depends on a lot of factors, chief among them your innate curiosity about your lineage and about your ancient patriline. Your Y STR results circumstances (see Part III about interpreting results), and the availability of the original STR tester to provide additional swab sample if needed could be other factors.
Many people rapidly lose interest in their Y STR results when they don't get fast answers that help them fill some slots in their family tree.
They frequently also lose interest if they get instant surname matches, have established contact with their matches, and decide there isn't more to learn from the DNA.
If your STR results show complications similar to those illustrated in Part III, a Y SNP test will teach you which STR matches to concentrate on and which to ignore.
If you are a historian of your surname (professional or hobbyist), and you want to see how the various patrilines fit in the context of the surname as a whole, Y SNP testing definitely provides a better evaluative framework.
-
ISOGG has great data on the haplogroups. They are lettered from A through T. If you click where it says Tree Trunk you'll see how these very ancient branches were once converged on "Adam", then gradually diverged.
Y haplogroup F in particular causes some difficulties and F carriers can be confused with other haplogroups. Pure branches of F have not been sufficiently studied.
-
YES. If you read about STR doppelgangers in Part III, you'll soon realize that the coincidence of the same SNP mutation (same value mutating to the same new value at the same loci) occurring in different haplogroups is possible. These coincidences occur with some regularity.
Here is an example of the same SNP mutation occurring in two different haplogroups:
I2a M223 > L801 > CS6433 > L1272 > Y5717
R1b M269 > DF13 > FGC5494 > BY7804 > BY11594/Y5717 -
FTDNA offers individual SNP tests, SNP packs, and the discovery test Big Y.
NatGeo transfers are also accepted. At this time, NatGeo results are not integrated into the yDNA Haplotree.
Individual SNPs
Pros
✔ The individual SNP test will give an instant yes/no answer as to whether you are positive for that SNP.
✔ If you are positive for that SNP and that SNP is younger than your last known terminal SNP (i.e, further along the haplotree), the new result will appear in your haplogroup badge. When your matches view your results in their STR results tables, they will see your new result.
✔ A haplogroup project administrator might be able to make educated guesses as to what individual SNPs you should test.
✔ This is a super low-risk option if the original tester is not available to do additional swabbing. There should be sufficient swab sample left at FTDNA to be able to finish the test successfully.
Cons
✘ If you are lost in the SNP wilderness with no hints from any STR matches about your branch in the Y haplotree, individual SNPs are not a good choice. See SNP Packs or Big Y instead.
✘ Your individual SNP result is not integrated into the FTDNA Y Haplotree.
✘ Does not complete your basic STR testing if you haven't already done so.
✘ FTDNA does not show you matches who have tested the same SNP, though some of them might be in your Y STR matches.
✘ FTDNA does not reevaluate your result for a possible terminal SNP downstream.
✘ At $39 per SNP at FTDNA, the individual SNP test is expensive. There are other labs that will test individual SNPs more affordably, but their results are not integrated into FTDNA's results.
✘ Individual SNP tests rarely go on sale.
✘ There is an EXTREMELY SMALL chance that even though your patriline carried a certain SNP, there was a copy error from your father to you so you might through that accident be negative, whereas your brother might be positive. Without knowing the context of your full SNP backbone, we won't know if this occurred.
✘ Once you've gotten one result, you might feel compelled to spend more money testing more individual SNPs. If you test three SNPs, you've already spent more money than if you had ordered an SNP pack, which is usually a better option.
SNP Packs
Pros
✔ If you are lost in the Y DNA wilderness, with no Y STR DNA matches with whom to compare, an SNP pack is an affordable option. Your predicted haplogroup is a good starting point. There is very likely a Y haplogroup project that covers your predicted haplogroup. The administrators are often able to provide further guidance on the merits or drawbacks of an SNP pack. Haplogroups Q, R1a, and R1b have backbone packs.
✔ Most SNP packs run $119 USD, and test roughly 100-200 SNPs. Compared to the individual SNP tests, this is a highly economical and affordable option. You are paying somewhere in the range of 60 cents - $1.19 per SNP tested, compared to $39 per individual SNP.
✔ If your SNP pack test successfully places you further along in the haplotree, your new terminal SNP result shows up in your haplogroup badge and in the matches table of your STR matches.
✔ This is a low-risk option if the original tester is not available to do additional swabbing. There should be sufficient swab sample left for FTDNA to test 100-200 SNPs and be able to finish the test successfully.
Cons
✘ The SNP pack is not a discovery test. Once you've tested those SNPs, your results will not undergo further refinement. In other words, FTDNA will not reevaluate your results for another terminal SNP downstream. You are simply testing a collection of SNPs without any continually updated analysis of the relationship of those SNPs to each other, which could change.
✘ Does not complete your basic STR testing if you haven't already done so.
✘ Your SNP results are not integrated into the FTDNA Y Haplotree.
✘ FTDNA does not show you matches who have tested the same SNPs, though some of them might be in your Y STR matches.
✘ SNP packs rarely go on sale.
✘ Some SNP packs do not probe deeply enough into the branch that could be applicable to you. You can click Add Ons & Upgrades in the top menu bar of your account dashboard and explore the SNP packs. Before purchasing an SNP pack, you should ask your haplogroup project administrator about the merits of the pack. If you see a lot of STR matches with a particular SNP, you'll probably want an SNP pack that covers that SNP. SNP packs are often limited in their coverage. There isn't an SNP pack for every region of the Y Haplotree.
✘ If you have fully tested out 111 markers then spend money on two SNP packs, you would probably have been better off just waiting for a Big Y sale.
Big Y
Pros
✔ Analyzes over 200,000 SNPs. This is a SNP discovery test, so your results, including your terminal SNP, will continue to be reevaluated. Consider it an automatic subscription for new information about your SNP branch.
✔ No worries about coverage of specific SNPs you see among your STR matches. They'll get covered.
✔ Completes your basic 111 STR marker testing if you haven't already done so.
✔ Whenever your terminal SNP is revised, it gets updated in your haplogroup badge and in the matches table of your STR matches.
✔ Reads several hundred more STR markers, an area of research not yet heavily explored.
✔ Your results are integrated into the FTDNA Y Haplotree if your account is set up for data sharing.
✔ There is a set of tools in FTDNA specifically for viewing Big Y matches.
✔ Though relatively expensive, if you've already completed Y111, $4.60 saved per week for a full year will get you Big Y, not considering further sale discounts.
✔ Is typically on sale over Father's Day and at the end of the year.
✔ You can order a raw data file of your genome from your Big Y result if you want to share your data with other projects (though there is an extra cost involved if you order Big Y after November 1, 2019).
Cons
✘ With no prior Y STR testing at FTDNA, the price is (currently, in 2020) $449 without sale discounts considered. The good news is, the price has been working its way down, and the more Y STRs you've tested prior to Big Y, the less it costs. Father's Day, mid-summer, and end of year sales are the best times to acquire the test.
✘ The most recent version of Big Y requires a lot of good swab sample. There is a moderate-to-high risk a reswab will be needed if the prior swab is old or was poorly or insufficiently taken. You won't know if the reswab will be needed until the test is attempted. Big Y is not recommended if the original tester is incapacitated or deceased or is unable to provide a good reswab without in-person guidance.
-
On your dashboard is a collection of widgets labeled Big Y.
-
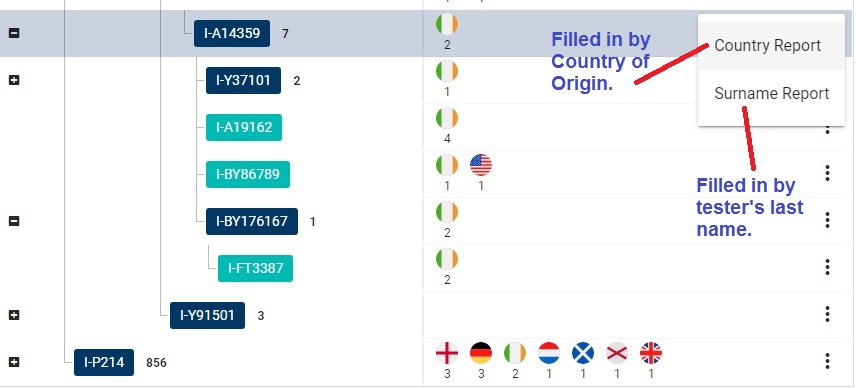
If you have taken the Big Y test, you can add your data to the database, thereby improving its quality:
✔ From your account settings, share out your DNA and ancestral origin results.
✔ Completely fill in your paternal origin data, including country and even GPS coordinates if possible.
✔ Make sure the Last Name Field of your Contact Information is clean and contains ONLY your last name. This is NOT the place to stuff your GEDMATCH number, phone number, etc.
When you pay attention to those details, the yDNA Haplotree can generate more accurate Surname and Country reports.
-
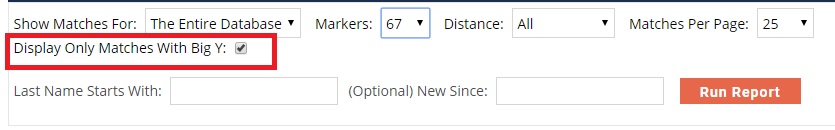
This option appears in the Y STR results search box ONLY if you have also tested Big Y.
The dialog box for filtering your Y STR matches now includes a checkbox that lets you view your Big Y tested matches.
-
YES. And the reasons are not entirely clear. Here are some ideas.
Assuming you and those matches are all enrolled in your surname project, your surname project administrator can look at the Genetic Distance separating you from your matches to find out the actual GD. The administrator can look more closely at the specific mutations causing the wide GD to determine the STR markers on which those mutations occurred. They might be on exceptionally volatile markers. Or maybe there is some unusual volatility on your Y lineage in general. If you assume 1 mutation per generation, and there are 3 generations per century, then it only takes 11 generations , or about 3 1/3 centuries back to a TMRCA, to push you and your match off each other's STR match lists. That is not so very long ago.
Two people are pushed off each other's STR match lists at 111 STR markers if their GD exceeds 10.
The terminal SNP that you share might be thought to be relatively recent, when it could be considerably older. Chances are your terminal SNPs will eventually undergo further evaluation, get revised time estimates, and you and the SNP match could end up in different subclades.
-
Yes. Recall our discussions in the earlier parts of this tutorial about DNA sometimes being imperfectly copied and not always possible to read.
-
At this time, SNP formation dates are not in the published Y haplotree. However, there are many other options.
👉 Y testers can click the Time Predictor Tool on a match to view the TMRCA table showing the time estimate for the tested pair.
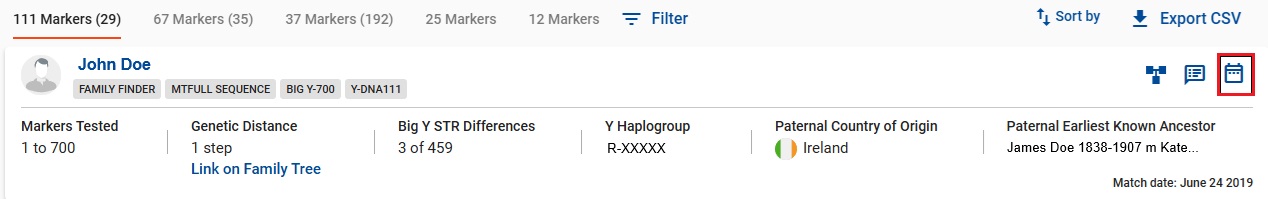
👉 From the Big Y menu, click on Discover Haplogroup Reports to see a rough timeline and history of your haplotree branch. To make this tool more valuable, Big Y testers should make sure Paternal Ancestry data is completely filled in under Account Settings | Genealogy, including country origin and GPS coordinates if possible.
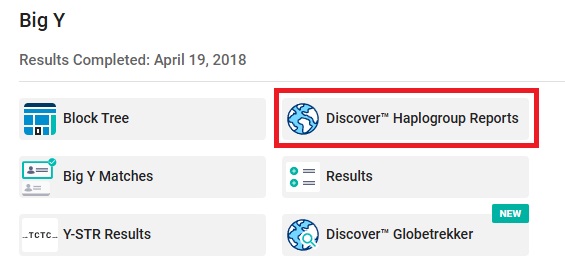
Within the Discover Haplogroup Report pages, you should be able to access many features such as Country Frequency, a Match Time Tree, and a Classic View of your haplotree.
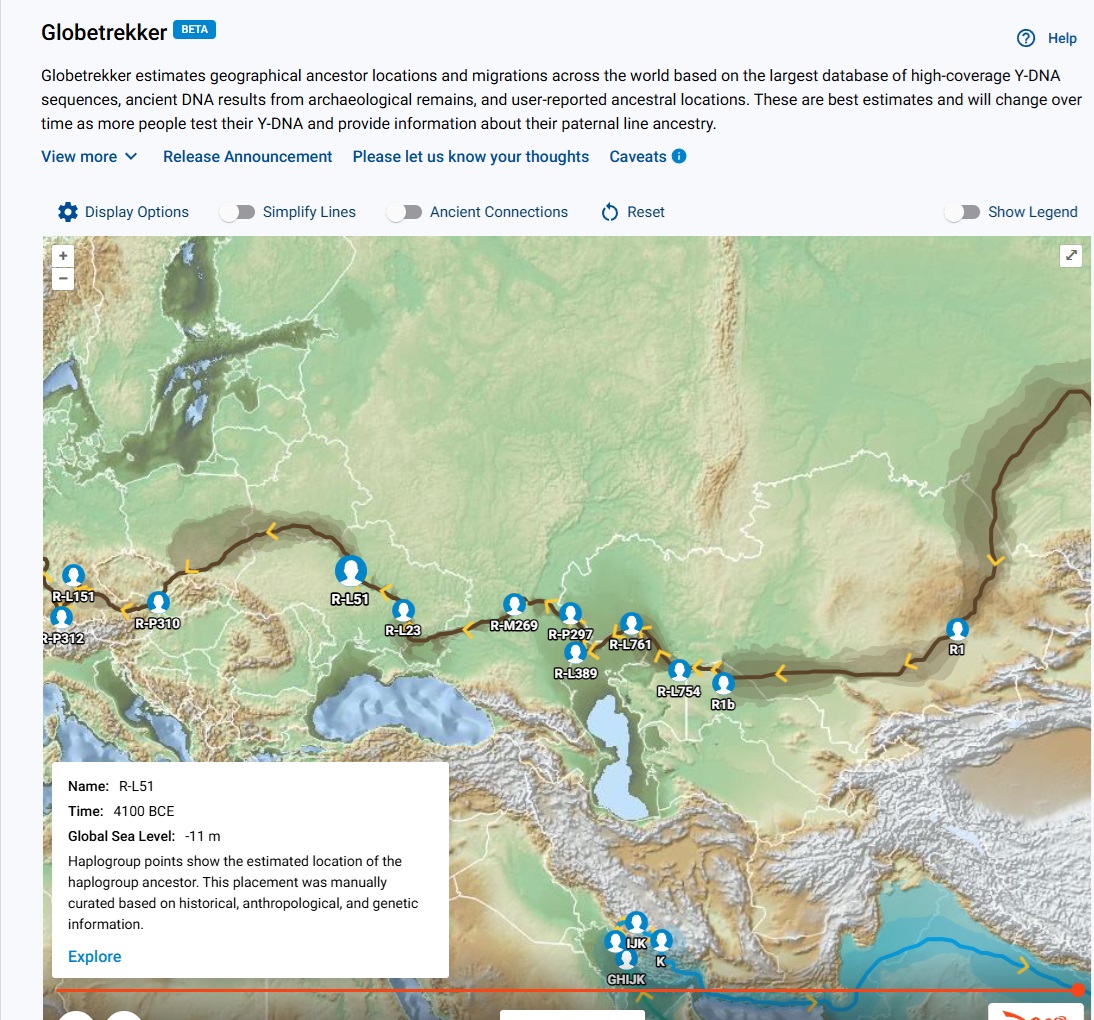
👉 From the Big Y menu, click on Discover Globetrekker to see a map of your haplogroup branch. You can click on an SNP on the map to see the TMRCA for that SNP.
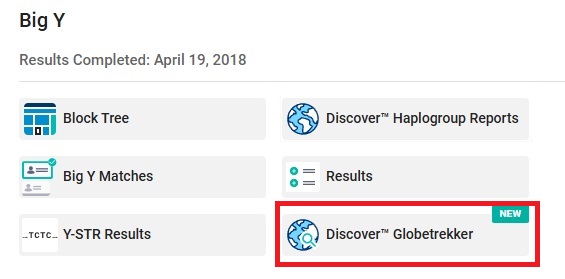
Within Discover Globetrekker, you can click on an SNP to display the SNP's approximate formation date or TMRCA.
👉 Some FTDNA projects also enable a Group Time tree.
👉 Some FTDNA projects also utilize SNP Time information from other websites.
-
A no-call is an attempt to read a location on the Y chromosome and a value cannot be determined. No-calls occur on both SNP and STR reads.
Besides SNPs, Big Y tests many hundreds of additional STRs, which contain many no-calls. Technology improvements and another attempt at reading those locations could reduce the number of no-calls.
-
Join a haplogroup project! You are probably eligible to join more than one, as there are numerous haplogroup projects representing SNPs in your SNP stream between the top of your haplotree and your terminal SNP. Your surname project administrator can help you find relevant ones. Your haplogroup project administrators will greatly appreciate your results.
Be aware that some haplogroup project administrators prefer that you have tested 67 markers or more because they like to look for correlations between STR marker mutations and SNP mutations. If you have done the Big Y test, your STR marker testing will have automatically been completed.
Your project might maintain a list of relevant haplogroup projects in the LINKS at the FTDNA website for the project. Ask your Y surname project administrator for the links to relevant haplogroup projects if you cannot find them.
-
FTDNA warns that the Big Y Matches tool can lead to confusion.
The matches tool lists Big Y testers who share a SNP mutation with you. You can search for matches by their name or by a mutation name (variant). These matches may or may not actually be on your terminal SNP branch. Furthermore, matches on your same terminal SNP branch may or may not share mutations with you along that SNP limb.
Matches sharing a mutation. Some matches show up because of the coincidence of a particular mutation occurring in men belonging to different haplogroups. Recall our discussion of SNP doppelgangers.
Matches sharing a terminal SNP branch but missing a mutation. Some matches in your SNP branch may fail to turn up when you search for certain variants (SNPs) along that branch. Recall the discussion under Missing Mutations.
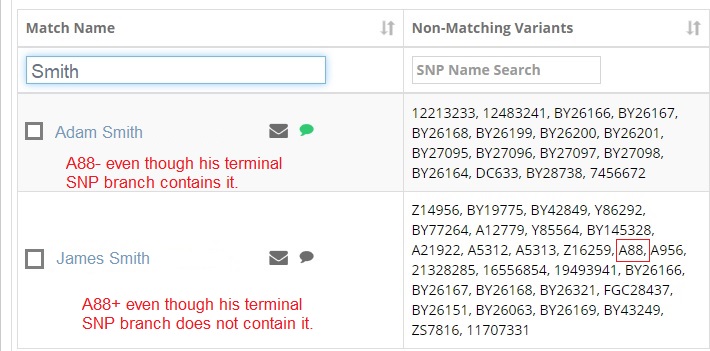
An anonymized real-life example is shown below. The tester, "John Smith", is searching for SNP matches with his last name. John Smith falls under R-CTS4466 ... A88 > Z16259.
John's first surname match below, "Adam Smith", is a second cousin sharing a paternal line ancestor. Adam's Big Y testing was problematic, requiring multiple swabs. There were problems reading his data. He is missing a positive read of A88.
The second match, "James Smith", happens to test positive for A88 but his terminal SNP is in a branch under R-CTS4466 that does NOT include it. James Smith is not a paternal line cousin to Adam and John.
It is coincidental that James shares the same surname as John and Adam.
The potential for great confusion here is probably why FTDNA steers you towards the Block Tree Tool (below).
-
This is a tool that gives Big Y testers a view of the cousin branches to their own particular SNP subclades that the normal Big Y SNP matches view does not provide. It gives a magnified blowup of the Y Haplotree in your part of the tree.
There are horizontal scroll arrows to help you navigate across the cousin branches. The branches are represented by blocks. Your own subclade block will have a black outline.
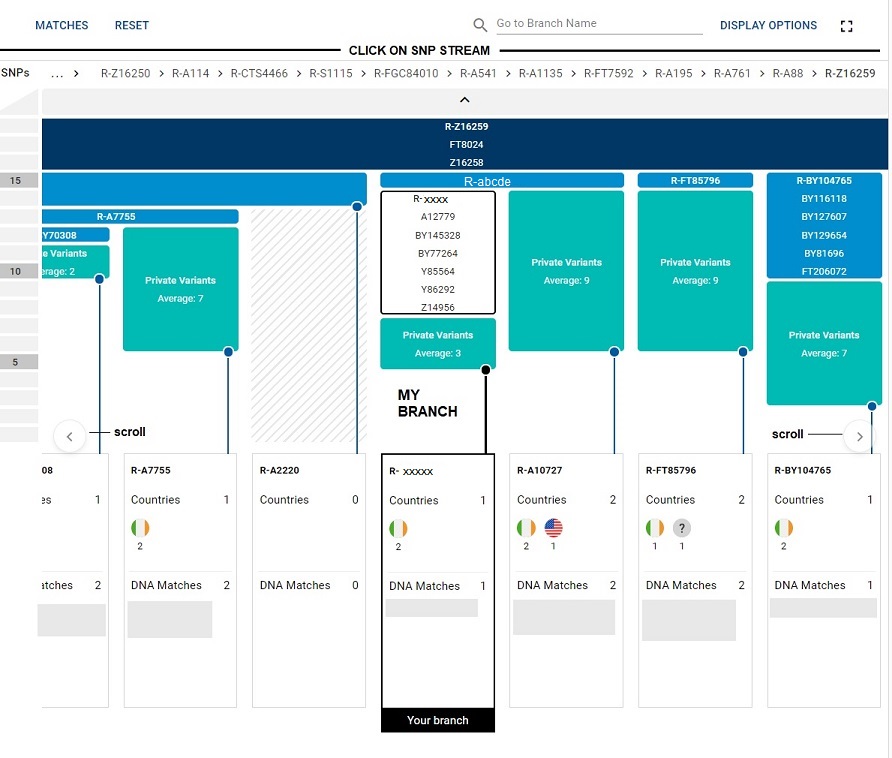
The branch blocks will list SNP matches in those blocks below where the block says Countries then DNA Matches. In the graphic the names of the actual SNP matches for this tester are grayed out.
At the top of the display is your SNP stream. You can click on an SNP in that stream and see the blocks below that SNP. If you get lost, click the RESET button and that will anchor the block tool back to you.
There is more detail on the Y-DNA Block Tree at the FTDNA Learning Center.
-
YES. Even though it is not the newest version of Big Y, Big Y 500 results are still continuing to be refined over time and the results are integrated into FTDNA's haplotree and matches.
-
See FTDNA's Rootstech video: Y-DNA: How SNPs Are Added to the Y Haplotree
There is more on the Y-DNA Haplotree at the FTDNA Learning Center.