Part III: Understanding and Navigating Basic Y STR Results
This page may be embedded in an HTML frame. If so, be sure to utilize the vertical scroll bar on the HTML frame as well as the vertical scroll bar in your browser. On the right side of the browser window, the frame scrollbar should be left of your browser scrollbar.
| marker | formation date |
| Short Tandem Repeat (STR) | Time to Most Recent Common Ancestor (TMRCA) |
| allele | Single Nucleotide Polymorphism (SNP) |
| haplotype | haplogroup and terminal SNP |
| modal haplotype | non-paternity event (NPE) |
| Genetic Distance (GD) | haplotree |
| advent of surname usage | clade and subclade |
| mutation rates | upstream and downstream |
Prerequisites: Part I and Part II are recommended.
- How does the yDNA test work?
- What is a marker?
- What is a Short Tandem Repeat (STR)?
- What is an SNP and what is a haplotree?
- What is a haplogroup?
- What will I get in my results with a basic Y STR test?
- Where can I access my Y STR results?
- What do my Y STR results look like in my FTDNA account?
- What do these numbers mean?!?
- What will my Y DNA results like on a Y results page?
- What is a haplotype?
- What is a modal haplotype?
- How are surnames relevant to my Y results?
- Will I have matches?
- How do I interpret my matches?
- Why don't I have Y matches?
- What do I do if I have no meaningful Y matches?
- What information is available about my matches?
- What is Genetic Distance (GD)?
- Are my closest relatives always next to me in the yDNA results tables?
- How far apart must another tester be before he is no longer a match?
- Do close matches prove my genealogical research?
- Why don't some of my matches show contact information?
- What is the FTDNA Time Predictor?
- How long is a generation in years?
- What is a mutation rate?
- How do the formation date and TMRCA of a mutation differ?
- Can Family Finder or other autosomal testing help with my Y DNA research?
- What is the relationship between my haplotype and my haplogroup?
- What is a clade and a subclade?
- What is a terminal SNP?
- How do I look in the FTDNA Y Haplotree?
- How can FTDNA make assumptions about my parent SNPs?
- What are the yDNA Haplogroup and Terminal SNP Columns in my Y Matches?
- Does a perfect 37/37 match prove we are related?
- Is is possible to have a perfect 111/111 match and not know how we are related?
- Is it possible to have a good match at 111 markers who is NOT related within a genealogical timeframe?
- How far back are STR matches reliable?
- How do I sort, search, and filter my Y STR matches?
- What is a palindromic multi-copy marker?
- Why should I upgrade my markers if I have few/no matches at 37 markers?
- Why are there so many null values in my Y111 results?
- If I want to upgrade my markers do I need to submit another cheek swab?
- My second cousin and I both have the same surname and have the same great grandfather yet we are in different haplogroups. I refuse to believe a non-paternal event occurred. Isn't it possible to be related yet in different haplogroups?
- I am descended from a paternal ancestor living 300+ years ago. Another person in a different major haplogroup and whom I do not match is claiming the same paternal line ancestor. The ancestry is well documented. How can this be?
- Where can I learn more?
-
The X sex chromosomes in a female and the autosomal chromosomes of either gender recombine to create one chromosome to pass to a child as part of the pair the child receives. The Y chromosome does not generally recombine the way the others do.
There is DNA in the non-combining part of the Y chromosome that almost never changes so is considered very stable. This DNA is used to identify the branches of the human family tree, going back many tens of thousands of years. In order to confirm your human family tree branch, you would have to get an advanced Y test. However, when you do a standard Y DNA test, FTDNA will attempt to predict the parent of your family tree branch.
This part of the Y chromosome at one time had been called "junk" because it was once believed that this part of the chromosome was obsolete and no longer had any life-sustaining functions for the man. In this region are locations that are points for comparison, somewhat analogous to comparison points in fingerprints. One point is called a locus; multiple points are called loci.
Two testers who have identical or nearly identical matching DNA at the comparison points probably share relatively recent common ancestry. The more comparison points tested, the greater the accuracy. The DNA fragments at these loci are scientifically studied to determine how often they change (mutate). Some mutate very quickly (every few generations) and some mutate quite slowly (maybe once or twice in a thousand years). Taking into account different mutation rates for DNA at different loci, the testing lab estimates how many generations back you and another tester share a common ancestor.
There are two types of Y-DNA that are interesting for genetic genealogy. The standard Y test analyzes loci called markers (see below). This test can help find recent genealogical matches when your results and that of another tester are identical or nearly identical.
Other Y tests identify the sequence of mutations starting from the dawn of man and occurring to the present day, forming your patriline backbone. Your recent genealogical matches, as determined by STR matching, must also carry the same SNPs in order to be true cousins. Much research is being done on SNPs, and they aren't just for identifying ancient branches any more.
👉 Historically men have passed both Y chromosomes and their surnames to their sons, so Y DNA and surnames have very good correlation. This is the focus of Y DNA surname projects.
👉 Women then cannot do a yDNA test. See Part I if this is not understood. If they are interested in patrilineal clanship they can get their husbands, brothers, fathers, uncles, male cousins etc to test.
There are two parts to your yDNA relevant to your genetic genealogy. The first part is your patriline backbone, all the way back to the dawn of man. The second part consists of certain mutations you share with your relatively recent cousins. The two parts work independently of each other, but together give the complete yDNA picture. The second part is where men typically start with the exploration of their Y genetic genealogy.
-
A marker is a point-of-comparison location on the Y chromosome. A number of markers along the Y chromosome have been identified as useful for ancestral testing. The more markers tested, the better the information we get from the test.
-
An STR is a marker that repeats a molecular DNA pattern.
Two men are an STR match at the same loci if they have the same number of copies of a molecular pattern at that loci.
The basic Y DNA test at FTDNA is a Y STR test.
-
SNP ("snip") means Single Nucleotide Polymorphism. Elsewhere on the Y chromosome are locations useful for determining your ancient paternal line ancestry up to the present day. These single mutations occur in a particular sequence over time that cause branching, developing a haplotree.
To be true cousins, your STR matches who are close relatives should carry just about all the same SNP mutations you carry.
Short names for SNPs are written with a letter followed by a dash followed by a short name for the SNP, for example R-M269. The first letter represents one of the main Y DNA phylotrees. Sometimes, when the context is understood, the initial letter and dash are omitted so you will see just the name, i.e., M269.
In time estimates for the points at which branches of the Y DNA haplotree developed, SNPs are considered far more stable and make better "clocks" than STRs.
-
A haplogroup is a paternal clan sharing a parent SNP mutation or sequence of SNP mutations, thus sharing a common ancestor. You might hear your deep SNP ancestry called your backbone.
-
You get two types of information in your results.

Haplogroup BadgeLook at your account dashboard in the right sidebar section called Badges. There you have a high-level ancient ancestry (haplogroup) prediction.
The major Y parent haplogroups all over the world can be seen in the index at ISOGG. The parent haplogroups that we might most often see in the predictions at FTDNA, because so many of its testers have a Eurasian patriline background, include but are not limited to: E-M2, E-V13, G-M201, I-M253, I-P37, I-M223, J-M172, R-M198, and R-M269. Occasionally we see others, such as F-M89. Haplogroup D is common around eastern Asia. Your predicted parent haplogroup mutation probably was formed many thousands of years ago.
If you took the Y37 test, you'll see your specific values on markers 1 through 37. Both are explained in much greater detail below.
-
Look at the dashboard in your FTDNA account. You'll see a collection of widgets in a group called Y-DNA.
-
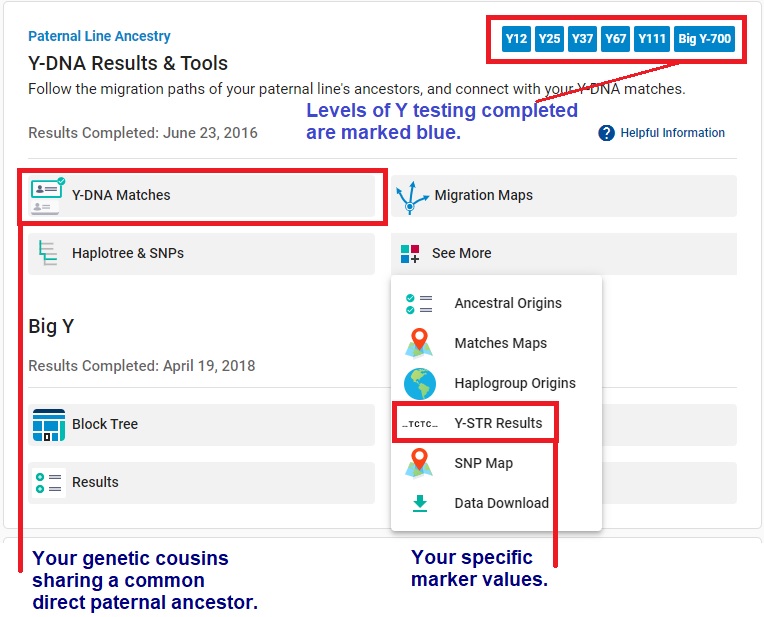
 Your STR values are listed in a table headed by the names of those STR markers, as in this example.
Your STR values are listed in a table headed by the names of those STR markers, as in this example.Results are divided into five panels, depending on how many markers you tested. Panel 1 covers markers 1-12, Panel 2 covers markers 13-25, Panel 3 covers markers 26-37, Panel 4 covers markers 38-67, and Panel 5 covers markers 68-111.
Markers are listed in a specific order. Each marker has a name associated with it.
Our usual minimal testing recommendation for new testers is 37 markers, which covers the first three panels.
If you click the little i icon next to the panel name, you'll get a FTDNA PDF file explaining your results in more detail.
-
Each number means that a certain molecular pattern repeated that number of times. For example, for marker DYS460 in the table above, the molecular pattern repeated 11 times.
👉 A marker takes a certain range of values. Each possible value for that marker is called an allele.
The lab finds your matches by comparing your values with others in their database. If two testers share an identical or near-identical pattern at 111 STR markers they are probably close genealogical cousins. This is never a 100% certainty.
-
If you are enrolled in a Y DNA project, your results and those of others are available in a table. The information you see about each tester may depend on what the administrator of your project has chosen to show. In the example here, each tester's row starts with a kit number (under the MIN MAX MODE rows); the actual kit numbers have been blanked out in the illustration. Next is information about the earliest known paternal ancestor if the tester has provided it. That information is also blanked out. Next is a predicted haplogroup (in red) or confirmed haplogroup (in green), and the STR values for that particular tester.
When you have project matches, the project administrator works to group you with your matches. An individual's results will then be juxtaposed to others in his cluster though not necessarily to his absolutely closest relatives.
Remember the collection of predicted haplogroups in the earlier question? If you see someone else in your project with a different parent haplogroup, it is immediately obvious that you do not share a genealogical patriline, even if his STR markers might look a little like yours. For example, if someone else's parent haplogroup starts with R and yours starts with I, you do not share a genealogical paternal ancestor.
-
Your haplotype is your specific sequence of STR values. This is also called your genetic signature. The table above shows the start of the genetic signatures for several people in predicted haplogroup E-M2. The sequence of numbers on the row of a particular tester constitute the haplotype of that tester. Most of the ones shown start with 15, 20, 17, 10, 15-17.
-
A modal haplotype is a haplotype created from a set of similar individual haplotypes by finding the most frequently occurring value of each marker. In the table above, the modal haplotype row is labeled MODE.
-
Surnames started coming into use late in the first millennium through the beginning of the second millennium. Roughly 900 C.E. - 1100 C.E. would be a good approximation. Keep this timeframe in mind as you evaluate the time estimates for the formation dates of your parent haplogroups compared to when your paternal lineage might have acquired its surname.
Surnames were more fluid up to relatively recently. Surnames may have been taken for protection, to hide from authorities, or they may have been awarded for services. Surnames may have changed because there were a lot of people in a small area with the same surname and one group of people wanted to distinguish themselves from others with the same surname. Some lineages may have acquired a surname relatively late, well after the aforementioned time window.
Surnames are more often than not polygenetic, coming from multiple sources. Not only do different genetic branches of man acquire the same surname but the surname itself could have come from different written sources. Different older written forms of the surname may have eventually merged into a single written form.
Here is an example of an anglicized name from very different sources. The name EASTMAN has been known to replace the Jewish EPSTEIN. It has also replaced the Scandinavian OSTMAN.
As you pick apart your Y results, you'll be comparing surname acquisition timeframes against haplogroup date timeframes.
-
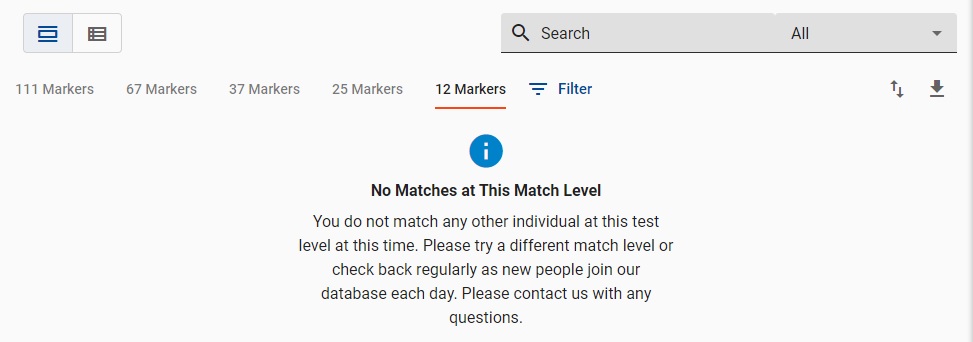
Very likely you will have at least one match.
It's rare, but a few men have ZERO matches. You're results display will look like this when that happens.
-
There are numerous possible scenarios into which your results will fall. Here are some of them.
CASE A. Let's deal with the most awkward situation first, a Non-Paternity Event (NPE) - also called misattributed paternity event. Suppose your paternal grandfather's brother's paternal grandson (your paternal line second cousin) has done a Y DNA test, and you decide to take one too. When your results come back, you do not match each other. Clearly, your paternal line pedigree says you should. You have uncovered a NPE.
It would take further investigation to figure out where the paternal line went astray. This could be the result of a hidden adoption, or a hushed up affair or rape that resulted in a pregnancy. If you had several qualifying male relatives in the extended family willing to test for you, it would probably not be too difficult to find out at what point this occurred. But this gets tricky for other reasons, because certain people in your extended family might be aware of the non-paternity and might be anxious to keep family skeletons in the closet.
Misattributed paternity events can be discovered by autosomal DNA too, which means that women can also discover their own misattributed paternity. And it happens more often that you'd think. There are Facebook groups devoted to shocking DNA results, and people discovering they are not who they think they are.
CASE B1. Now let's look at the most ideal outcome with your Y results. This is the outcome people typically expect and what most of us dream for. You have matches with your surname. Maybe you have LOTS of surname matches. You have probably found some relatives. You will have to work out how far back the common ancestor goes, genealogically. If your surname matches range from close to remote and you all have the curiosity and interest, you and your matches may want to consider advanced SNP testing to help carve out the branching of your family tree during the second millennium.
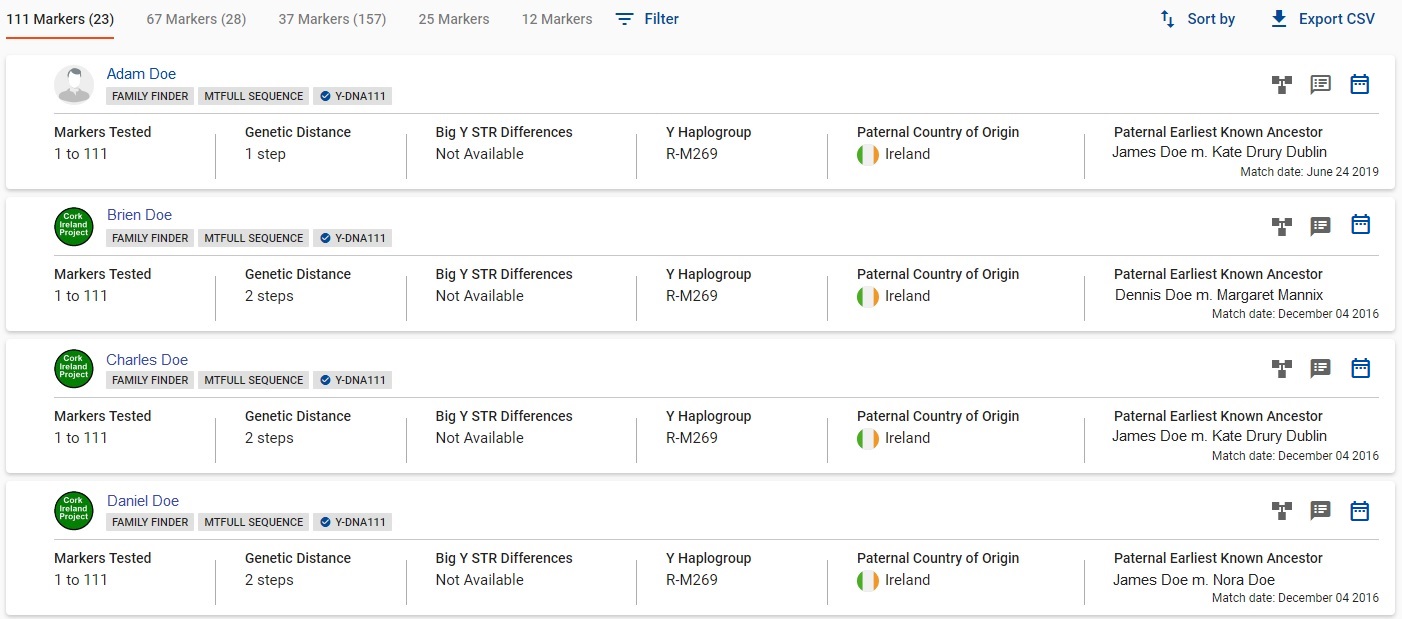
CASE B1: Almost ideal match situation.The illustration immediately above is taken from real data, with the surname replaced by DOE and the earliest known ancestor data replaced. It illustrates a very good outcome. It is "almost" ideal, because the DOE tester did not immediately have these matches upon initially receiving his Y results. A DOE relative had to do some research in the ancestral area and found four DOE men to test and confirm they share paternal ancestry. Though highly satisfactory, the situation is not quite 100% satisfactory because the lack of genealogical records in the ancestral area for the pertinent time frame (prior to the late 1810's) makes it impossible to identify the common DOE ancestor. These five testers don't exactly know how they are related.
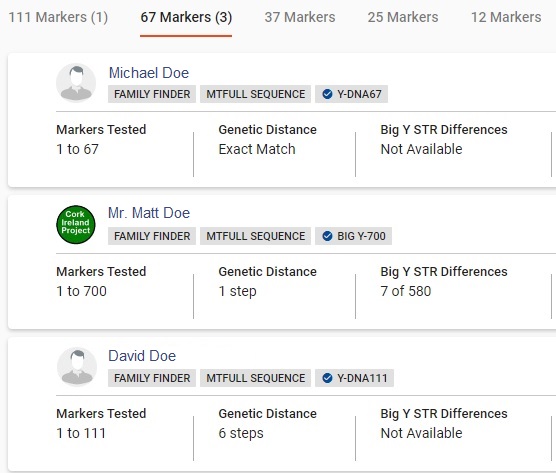
CASE B2: Polygenetic Surname MatchesCASE B2. Be careful interpreting distant matches if your surname is a very common one and widespread. It is very likely polygenetic. It is possible for the surname to be acquired by men in different haplogroups. It is even possible for the surname to be acquired by men with the same late first millennium parent haplogroup that split apart. The different child branches could have acquired the same surname independently.
The Y results in the CASE B2 table are taken from real data, with a very common surname replaced by DOE. This is different data from the ideal match data in CASE B1 table. Here, the results are filtered by the original surname. TESTER DOE is known to be a third cousin to Michael Doe and a second cousin to Mr. Matt Doe. They are unrelated to David Doe, who happens to have the same surname but does not match Tester Doe or Matt Doe at 111 markers. In this case Tester Doe, Michael Doe and Matt Doe are in one subclade of R-CTS4466 and David Doe is probably in R-CTS4466 in a different subclade. The split between the two subclades likely occurred long enough ago to precede acquisition of the DOE surname.
CASE C: Consistent name matches not tester's surnameCASE C. You have matches, and a lot of them may share the same surname but it is not your surname. Interpretation depends on how close those matches are.
If they are remote matches, there is a possibility their paternal line branch split off from yours prior to the advent of surnames, and few or no people in your branch are tested.
If they are close matches, that could be an indication that you have a paternal ancestor who changed his name.
There is some similarity to this situation to CASE A (misattributed paternity). Your next step would be to go look up that paternal line second cousin and get him to test too, to confirm there was no such event in your recent family history.
The CASE C table is taken from real unfiltered data. These are the ONLY matches this tester has. Names of matches have been changed but all have the same surname. It is NOT the tester's surname. The tester has upgraded his markers to Y111 to see if the matches persist, to make a better determination as to whether the common paternal ancestor existed pre-start-of-surnames or post-start-of-surnames. Some matches persist at 111 markers, and they are a further distance. The data right now leans towards pre-start. The paternal line genealogy research has been hitting difficulties, but is still ongoing.
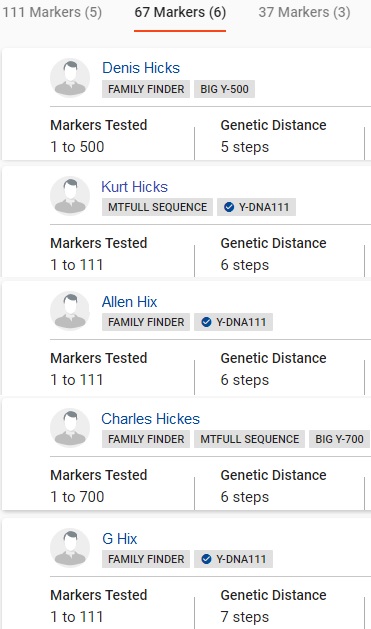
Of course, if your name happens to be HICKS, you've potentially hit the match jackpot (CASE B1).
The best interpretation here is that several men named HICKS (or spelling variation) tested and probaly shared a common paternal ancestor many centuries ago. Without running estimates, it's difficult to say how far back, but 8 centuries might be a good guess.
The lack of surname matches for the tester does not necessarily prove his paternal lineage has gone extinct. Neither does the abundance of HICKS matches. It is possible the tester's lineage has gone extinct, but one cannot tell by the given data.
CASE D. You have matches, but they come in a variety of surnames. In this case your data is NOISY. Upgrading your markers to Y67 or Y111 can help eliminate some noise. But even at 111 markers, data can be noisy, and the most recent common ancestor may have lived prior to surname acquisition. Sometimes Y SNP testing brings further clarification. You may find yourself falling into situation E.
CASE E. You may have NO matches or NO matches that are meaningful to you. Whatever matches there are have not even done any advanced SNP testing. Your data is very QUIET.
Even in CASE C, there was SOME information - that you and several men named HICKS shared a common ancestor. You can look at the haplogroups of those matches and maybe learn a little more. But here in CASE E, you don't even have that benefit.
Y SNP testing will clarify where you fall in the Y haplotree, but it is not likely to help you fill in your family tree back a few generations. Either some hard-boiled genealogy detective work might be what you need to break the research impasse. Or, you might have to passively accept the situation and wait and see if any meaningful results materialize.
********
There are some good messy examples further down this tutorial. See: What are the yDNA Haplogroup and Terminal SNP Columns in my Y Matches?
-
A. Men in your paternal line branch have not Y STR tested or their results are not in the FTDNA database.
B. Your patrilineage is going extinct. Yes, lineages of humans go extinct, just like other animals.
-
Try to find another eligible male relative who also has a direct paternal line of descent from your direct paternal ancestor (2nd, 3rd, 4th cousin, etc) and have him Y test.
If no such relative is conveniently available, you may have to look for men with your surname in the geographic area of your paternal line origins and make inquiries. In other words, really power up the genealogy research and aggressively search for such men. If you successfully end up finding, testing, and matching such a man, you've confirmed that at least as far back as your most recent shared direct paternal ancestor, there has been no non-paternity event. Your lack of matches could then be a case of relatives simply having not tested.
Otherwise you will have to resort to other measures, such as autosomal testing of as many relatives as possible - preferably in older generations - who share the same paternal line ancestry.
-
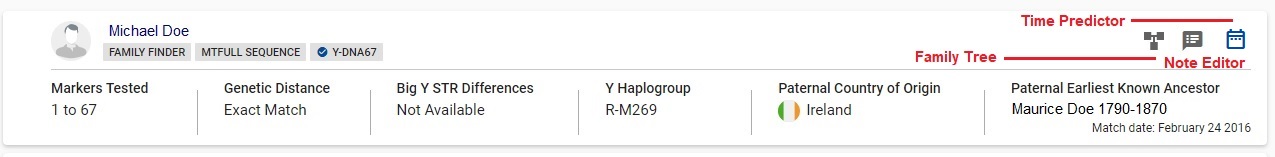
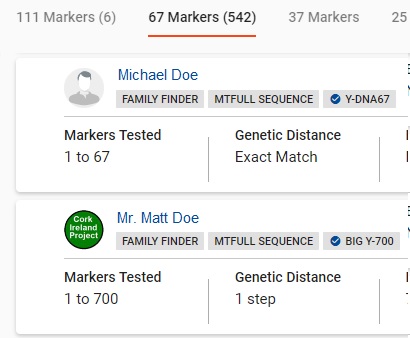
The Y results table Detail View shows the name of the match. Underneath his name are the DNA tests he has taken. In this example, Michael Doe has taken Family Finder, mtFull, and Y67.
Clicking on the match's name will pop open a window with contact email. If the match has entered Earliest Known Ancestors, that information will be listed. Next are the predicted or confirmed Y haplogroup, and the mtDNA haplogroup if that information is known.
The additional detail columns are: 1- Markers Tested; 2 - Genetic Distance; 3 - Big Y STR Differences; 4 - Y Haplogroup; 5 - Paternal Country of Origin; and 6 - Paternal Earliest Known Ancestor. The data in 5 and 6 are entered by the match.
There is a collection of utilities at the far right. If the match has entered in anything in a family tree, the Family Tree icon will appear dark gray, otherwise it will appear hollow. The second utility is a Note Editor for you to maintain notes about the match. The third utility is a Time Predictor back to a most recent common ancestor (formerly called TIP).
-
 Genetic Distance (GD) is the number of mutations between you and another tester. GD does not consider the stability or volatility of the markers on which those particular mutations occurred when you compare your results to others.
Genetic Distance (GD) is the number of mutations between you and another tester. GD does not consider the stability or volatility of the markers on which those particular mutations occurred when you compare your results to others.It is possible you could be more closely related to someone with whom you have a greater GD. The example on the right is taken from a real case. The first match, an Exact Match at 67 markers (GD = 0), is a third cousin to the tester. The second match, 1 Step from the tester (GD = 1), is a second cousin to the tester.
See: FTDNA's Measures of Relatedness.
This article from 2016 describes how FTDNA calculates GD.
Mutations occur randomly. A marker does not necessarily mutate in a particular direction, i.e., increasing or decreasing values. Markers can back-mutate, which we may not be able to detect. Nor do they necessarily mutate in single-step fashion; a marker can "skip" a value. It is possible for several mutations to occur in a single generation.
-
NO. It is OFTEN the case that our results will be juxtaposed to those of our closest relatives. But not necessarily so. One of the reasons was addressed in the preceding question. GD is a very general rule of thumb for determining how far removed you are from a match. But sometimes GD can mislead us. It is possible to be more mutations apart from closer relatives than you are from more distant relatives.
The results in yDNA results tables are likely character-sorted in ascending order from left to right.
Most of the time, the character sort of numbers would coincide with a numeric sort. The problem with the sort is the handling of the value 10. Numerically, the value 10 would sort after 9. But the character "1" in "10" would sort before the character "2", "3", and so forth all the way to "9."
Since the sort of characters "1" through "9" would coincide with a numeric sort of those numbers, "10" would sort before "11", and so forth.
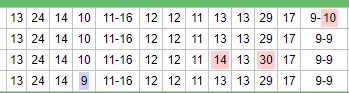 In the example shown, the first row shows the values 9-10 for the last marker shown, then the second row shows the values 9-9 for that same marker. If this were a numeric sort those two rows would have been reversed. Similarly, the last row shows a 9 at the fourth marker when all the rows above show 10. Had this been a numeric sort, the last row would appear first.
In the example shown, the first row shows the values 9-10 for the last marker shown, then the second row shows the values 9-9 for that same marker. If this were a numeric sort those two rows would have been reversed. Similarly, the last row shows a 9 at the fourth marker when all the rows above show 10. Had this been a numeric sort, the last row would appear first.Table sorting; results groupings; the number of your relatives who have tested and are grouped with you; how extensively you and your relatives have tested; and the nature of your mutations that sets you apart from your relatives will all affect how you are positioned compared to your relatives.
-
FTDNA publishes an Expected Relationships table. FTDNA uses specific Genetic Distance numbers to determine whether or not to cut other testers out of your match list.
These cutoff points for how closely someone is related to you are somewhat arbitrary. For example, third cousin patriline matches could be perfect 111/111 matches, and therefore fall into the "Very Tightly Related" category. Second cousin patriline matches could be 108/111 matches, and therefore fall into the "Related" category. But 2C cousins are closer than 3C cousins. So some common sense has to be applied to these guidelines.
See also a later question about upgrading markers from Y37.
-
In the GD example two questions ago, a great deal of genealogical research was done linking the tester and his two shown surname matches. While the Y test results don't unequivocally prove the exact degree of relationship between these three men, they do show that they all shared a relatively recent direct paternal line ancestor (d. 1867), giving powerful support to the genealogical family tree model on paper.
-
Some testers have set the match notifications in their accounts so that they are not participating at a particular level of testing. Therefore they will not share communication details with matches at that level.
-
The Time Predictor estimates how far back you and a match share a Most Recent Common Ancestor (MRCA). It is one of the utilities available in your matches table.
👉 The estimate calculated is called Time to Most Recent Common Ancestor, or TMRCA and used to be expressed in generations. On external project haplogroup pages for certain projects you might still see it expressed that way. The genetic differences on your STRs between you and those of your match are used to estimate how many generations back your most recent common ancestor lived. This is more than just a Genetic Distance count. The lab's estimate takes into account the average mutation rates it has of those particular markers that differ and their significance. For example, some markers are considered to be volatile, with high rates of change. Between known relatives we would downplay their significance. Slow-mutating markers probably have more significance. The slower mutations could be the more obvious indicators of branches of a paternal line. The mutation rates of particular markers may differ in different haplogroups.
If you've got two matches the same Genetic Distance apart from you, TMRCA results could be very different, depending on the STR mutations in play.
Here are illustrations of the older way of presentation.
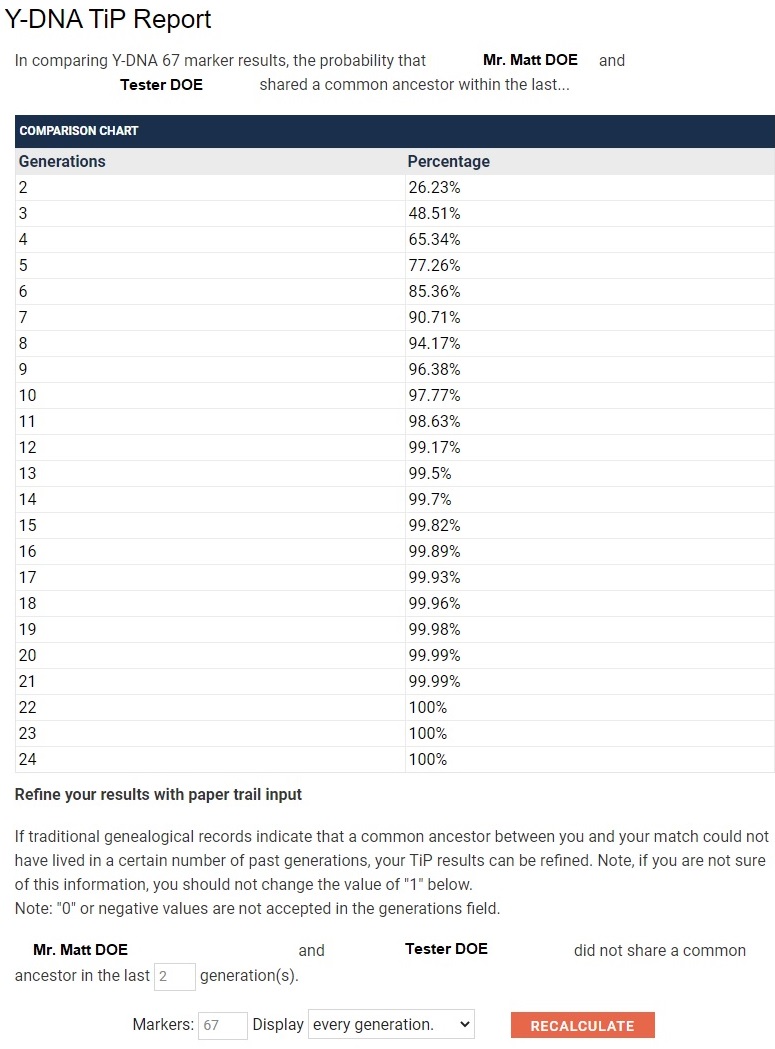
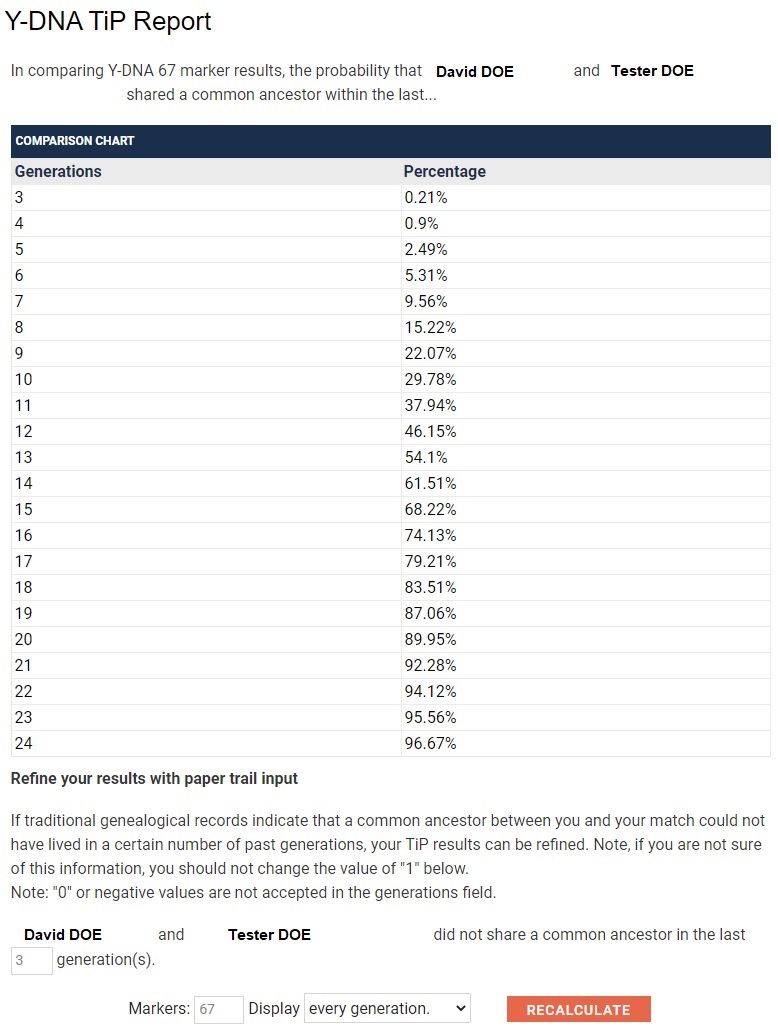
With data in such a form, you can chart and compare matches visually. In the following example, Tester A has two matches, both Genetic Distance 12. Tester A can chart the generations removed percentages to show that B is a closer match than C. For instance, you can instantly see that B crosses the 50% probability mark at about 11 generations, whereas C crosses the 50% probability mark at about 15 generations.
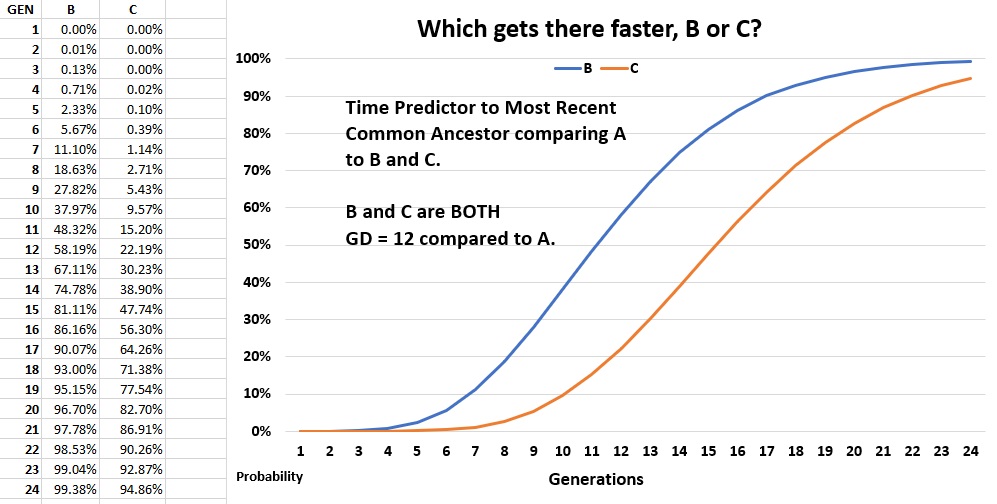
Time estimate calculations using STR mutation rates are increasingly unreliable the further we go back in time. Mutation rates used in TMRCA calculations and measures of relatedness are a set of averages. None of us are average. Mutation behavior can vary widely and change over time, randomly and unpredictably. While it would be rare, a father and son might show 3 or 4 mutations between them. Equally uncommon would be two men be 111/111 matches and not know their common ancestor going back several hundred years.
Even if our time estimate calculations are increasingly unreliable more than a few hundred years back, we might still use information about the relative stability or volatility of certain markers to consider the significance of a mutation between two matches. For example, suppose two matches differ from each other by mutations on markers considered volatile, but they also show a shared mutation compared to a modal haplotype on a marker that is considered extremely stable (very low mutation rate). The common value on a very stable marker could be a clue that the two matches should be carved out in the same branch.
STR markers have no memory of mutating, so their direction of mutation can go up or down.
FTDNA changed its presentation of TMRCA. Rather than present generations, it presents years. The sample Time Predictor table below shows Genetic Distance in the first column and the level of testing in the top row. The values in the cells are year ranges rather than generations. The name at the left on the bottom is the tester, and the name on the right is the match the tester is looking up. In this example, the tester and his match are Genetic Distance of 1 at 111 markers. Going across to the Y-111 TMRCA column, the TMRCA is 1850 CE, with a possible range of 1700-1900. Notice that the TMRCA is oriented towards the end of that range rather than the beginning. The TMRCA is not at the center of the range. The true TMRCA for these two real life testers probably preceeds 1800.
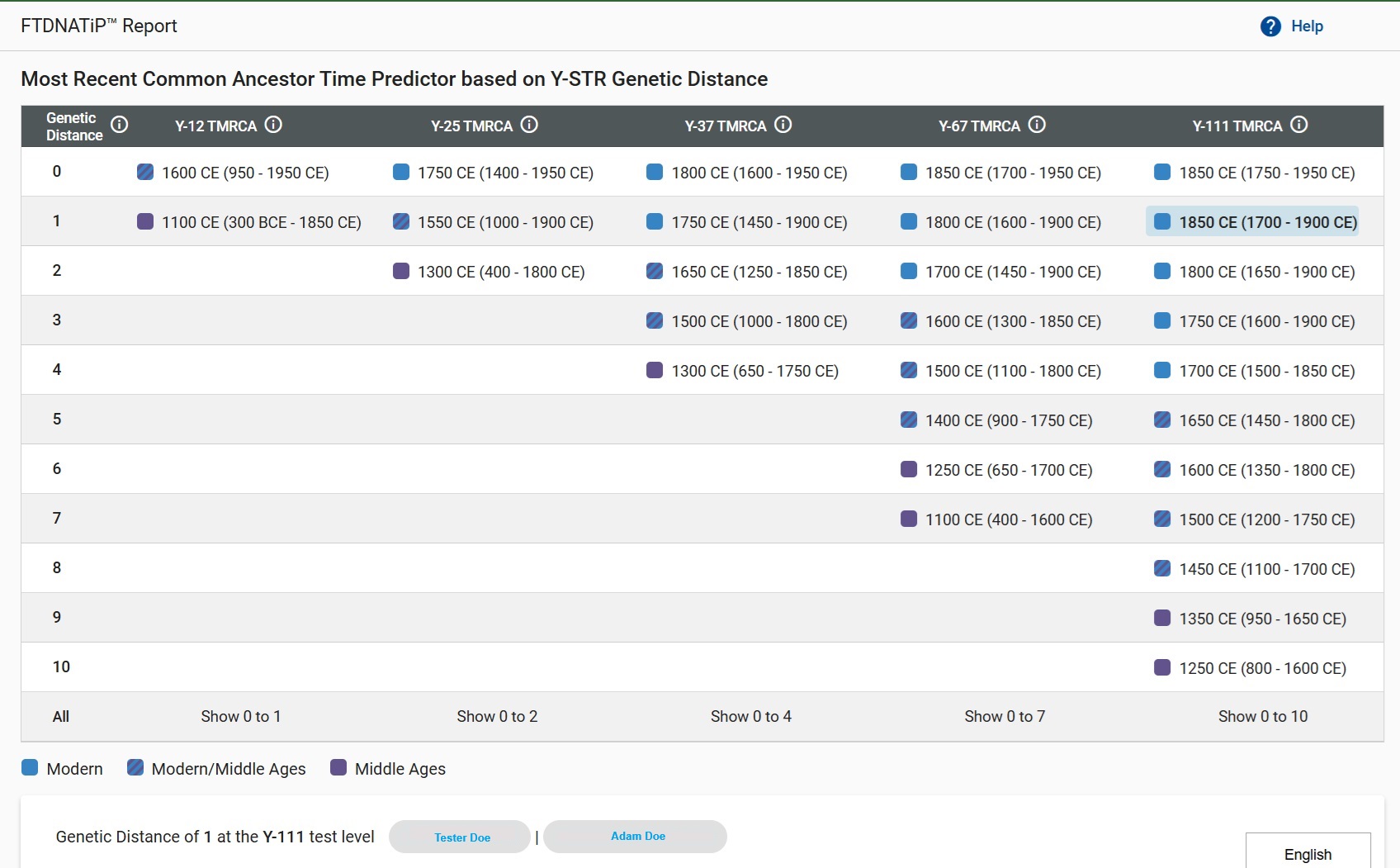
FTDNA now uses an additive model that considers both STR mutation rates as well as SNP haplotree point estimates.
-
An average number of 31 years per generation is consistent with actual pedigrees except that a wide range of values are observed as illustrated in the Sorenson database. They examined 131,758 father/son pairs and observed values as low as 12 and as high as 70. A TMRCA computation expressed as 3-20 generations prior is more accurate than multiplying by 31 and saying it was 93-630 years prior simply because whatever generation length is used it is a typical number that can be in error in and of itself.
-
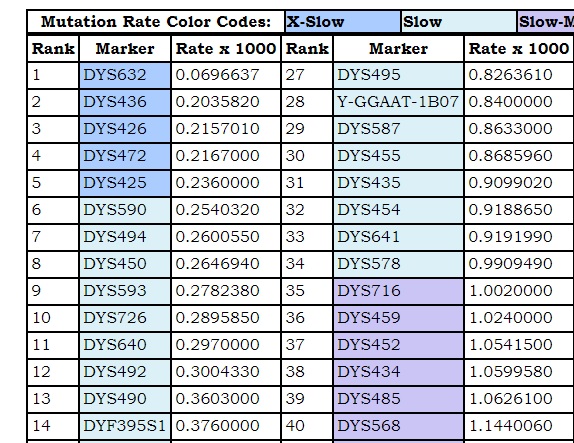
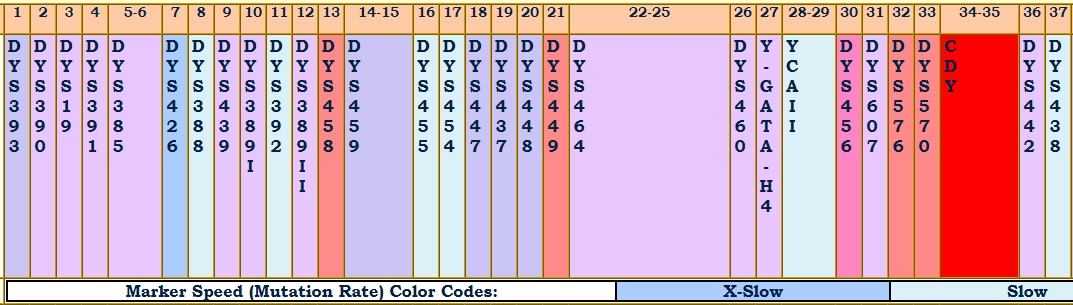
The mutation rate of a marker is a measure of how many times its value changes in a paternal lineage in a unit of time. The unit of time is usually a generation. Several researchers have come up with mutation rate sets and have ranked markers from those exhibiting the most stability to those with the greatest volatility. Since the numbers are so small, some tables show the mutation rate multiplied by 1000.
The image shows a part of a table of mutation rates. The mutation rates are ranked X-Slow, Slow, Slow-Medium, Medium, Fast-Medium, Fast, and X-Fast. The cutoff points for these relative rates were decided somewhat arbitrarily by the project administrator. To better illustrate those relative rates, the admin also assigned color codes, shifting from dark blue, through light blue, then bluish purple, then reddish purple, then red.
On haplogroup pages with haplotype tables, you will see the marker labels color-coded according to the markers' relative stability or volatility.
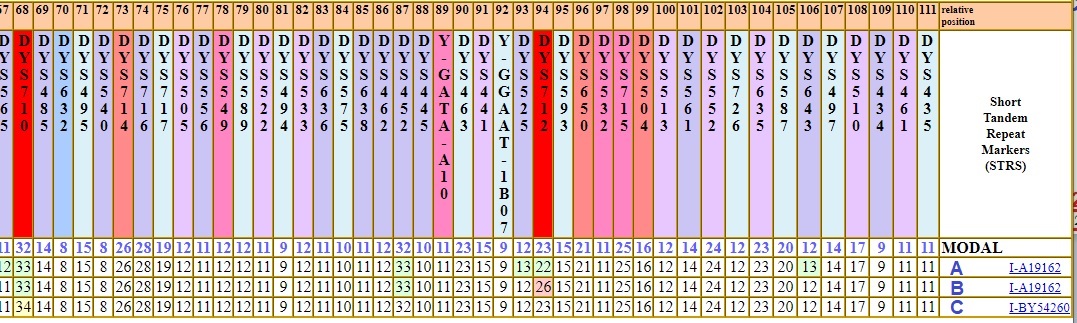
Mutation rates will matter when you are comparing matches and their Genetic Distances to you. Suppose you have two matches who are the same Genetic Distance apart from you, how can you tell which match is a closer relative? Here is one example.
Here, Tester A, is equal Genetic Distance from Tester B and Tester C (GD = 12.) Inside FTDNA, they would not show up in each other's Y STR match lists.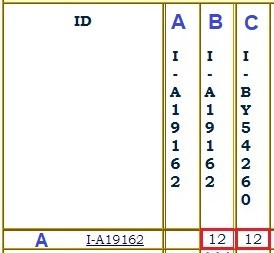
It so happens that all three testers did advanced SNP testing and we know what subclades they fall into. A and B fall into the same subclade, but suppose we didn't have that SNP information. Is there a way to tell from the STR markers who is likely the closer match?
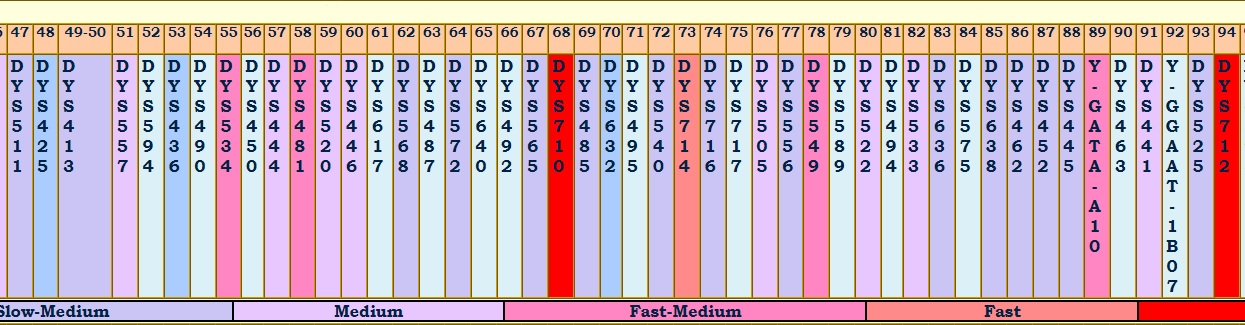
Let's take a look at the markers. In the partial haplotype table below, the modal is the first row, and is taken from the large group under which A, B, and C fall. Look at marker DYS712 in column 94. It is color coded bright red, indicating its relatively high mutation rate and extreme volatility. Notice that tester B is 3 steps different off the modal and 4 steps different from tester A. This one marker alone explains 4 steps of the 12 step difference between A and B. Furthermore, we don't really know if that huge jump occurred all in one generation or over several generations.
On that basis alone, one could argue (without the additional benefit of having terminal SNP data) that B is closer to A than C. The differences between A and C are far less "dramatic", showing gradual drifting apart over time.
If that is still not clear, the project's Time Predictor data based on STR mutation rates can be charted and compared visually. Here is another way of showing that B is a better match than C. For instance, you can instantly see that B crosses the 50% probability mark at about 11 generations, whereas C crosses the 50% probability mark at about 15 generations.

Time estimate calculations using STR mutation rates are increasingly unreliable the further we go back in time. Mutation rates used in TMRCA calculations and measures of relatedness are a set of averages. None of us are average. Mutation behavior can vary widely and change over time, randomly and unpredictably. While it would be rare, a father and son might show 3 or 4 mutations between them. Equally uncommon would be two men could have the same surname and be 111/111 matches and not know their common ancestor going back several hundred years.
Even if our time estimate calculations are increasingly unreliable more than a few hundred years back, we might still use information about the relative stability or volatility of certain markers to consider the significance of a mutation between two matches. For example, suppose two matches differ from each other by mutations on markers considered volatile, but they also show a shared mutation compared to a modal haplotype on a marker that is considered extremely stable (very low mutation rate). The common value on a very stable marker could be a clue that the two matches should be carved out in the same branch.
STR markers have no memory of mutating, so their direction of mutation can go up or down.
-
Imagine an ancient clan of men. They shared a common paternal line ancestor. During that time, one of them develops a new mutation and sires at least one son. The clan experiences a disaster, during which many of these men die, including almost all the men carrying the new mutation. The clan struggles to stay alive during the crisis, which lasts for many hundreds of years - say 20 generations. The number of men carrying the new mutation dwindles to just a single lineage. Finally, the clan migrates to a new place where there is an opportunity to rapidly expand its population. The lineage with the new mutation then propagates. You and your matches inherited that mutation.
Your most recent common ancestor will be dated AFTER the crisis. The formation date of that mutation was BEFORE the crisis. There can be a considerable gap of time between the two.
This is applicable to both STR and SNP mutations.
-
The short answer is YES. If you have a close yDNA match who has also tested Family Finder, consider also incorporating autoDNA in your investigation. If you and your Y match are also autosomal matches, that *could be* indicative of a relatively recent common paternal ancestor.
 👉 If your Y match shows up in your autosomal matches, you could be related to him in more than one way. This can happen more often than you'd think, for instance if your ancestors lived in the same close-knit rural community over several generations. You would need to rule out a multiple relationship possibility to conclude the autosomes are showing a match ONLY because of the relationship through the direct paternal line.
👉 If your Y match shows up in your autosomal matches, you could be related to him in more than one way. This can happen more often than you'd think, for instance if your ancestors lived in the same close-knit rural community over several generations. You would need to rule out a multiple relationship possibility to conclude the autosomes are showing a match ONLY because of the relationship through the direct paternal line.Whether or not you and your Y match are also autosomal matches, you should ask your Y match if he has had other relatives descended from his paternal ancestor also autosomally tested. Then you can search your autosomal matches for those names. In turn you should autosomally test lots of relatives also descended from your paternal ancestor, so your Y match can likewise sift through HIS autosomal matches for their names.
-
The short answer is that there is no causal relationship. Your SNP backbone determines your clan. Your STR mutations occur independently. STR mutations do not cause SNP mutations and vice versa.
Your genetic signature is your particular set of values from your Y STR test. A modal haplotype is a haplotype derived from the most frequently occurring values occurring in a collection of haplotypes. When we find testers with similar genetic signatures we attempt to put them together into such collections because they are showing evidence of being part of the same lineage.
A haplogroup of men share a common deep ancestry, as evidenced by SNP mutations. Usually - but not always 100% of the time - their STR genetic signatures resemble each other.
Sometimes - though not always - SNP mutations correlate with certain STR mutations. Research is strongly oriented towards finding SNP mutations (haplogroups) that correlate to particular STR-haplotypes, because we love the convenience of having STR predictors when we cannot do a Y SNP test.
As an example, we once spoke of Irish Type II, the South Irish modal haplotype, which possesses certain recognizable values. This genetic signature has a good correlation with R-CTS4466, which underwent rapid expansion starting about 200 C.E. So now we refer to the CTS4466 haplogroup.
The illustration below shows the genetic signatures of several men with paternal ancestry from the same location and with the same last name. This data would constitute the leaves on the trees in the pictures above.
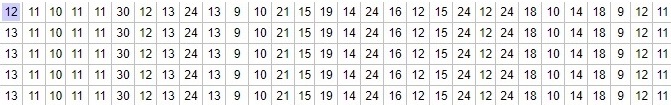
Partial Genetic Signatures of several closely related menThey fall under the R1b subclade R-CTS4466. Only one of these men has done advanced Y testing to further identify and refine their SNP subclade. They fall under:
R-CTS4466 > S1115 > FGC84010 > A541 > A1135 > BY19775 > A956 > A2288 > A5313 > A5312
If this were represented on the trees above, we would probably label the bottom of the trunk R-CTS4466. We could go back further, all the way to R, if you want to go back many many thousands of years. But R-CTS4466 is a good place to start since it is such an important SNP from the historical perspective of these particular men. From there, we'd move up the trunk, and out onto a branch, labeling S1115, FGC64010, A541, etc. The branch will get narrower and narrower as we progress towards the leaves.
Notice that as we label the SNP stream, the first SNP starts with R-. After the first SNP, we can drop the R- prefix since it is understood the SNP stream is under haplogroup R.
This labeling will be shown a few questions down (below).
-
A clade is a branch off the main trunk of the haplotree, and a subclade is a thinner branch off a thicker branch. Branches and limbs represent your deep ancestry, as determined by SNPs. The thicker the branch, the older that mutation. The relationship of a subclade to a clade is relative.
When considering a haplotree, we expect haplogroup and subclade to represent the same thing, in terms of junctions at which branches occur.
For example, R1a and R1b are different subclades of R, a major phylogenetic group. R-M512 is a subclade off of R1a and R-CTS4466 is a subclade of R1b. If your Y results show you are in R1a, you will not find your recent paternal line cousins over in R1b. Your recent cousins must share a relatively new branch with you under R1a.
-
A terminal SNP is the most recent SNP mutation defining the known end branch of your SNP backbone. FTDNA's awareness of your terminal SNP depends on how much Y SNP testing you have done.
The replication of yDNA between father and son is not always perfect. Sometimes the copy process misses something, or sometimes the lab has difficulty reading the value at a certain location. In rare instances, there are copying or reading errors when Y DNA is passed on. So sometimes you and your closest matches may not show the exact same predicted haplogroup or terminal SNP.
Even a father and son could have different terminal SNPs. The son could spawn a new mutation and thus have a new terminal SNP that his father does not have.
-
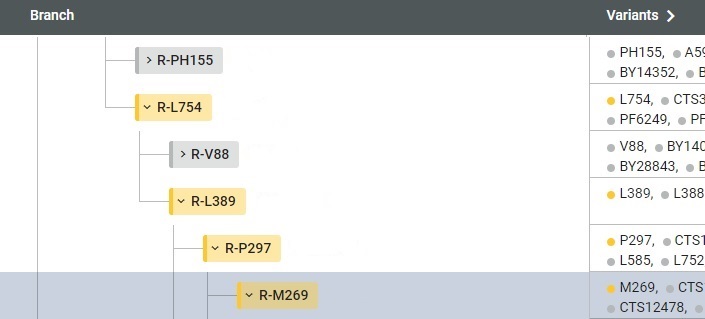
Click your haplogroup badge to see your position in FTDNA's Y Haplotree. The position of your haplogroup is highlighted with the gray bar. If you have done some Y STR testing but have not done any Y SNP testing, your haplogroup is a prediction.
-
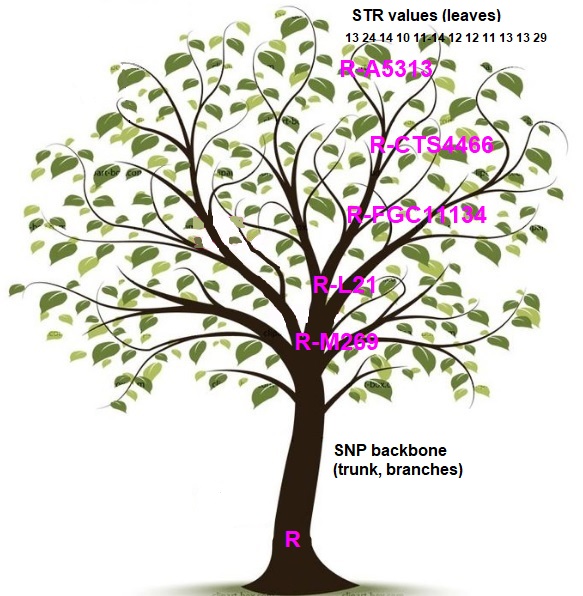
FTDNA can make assumptions because SNP haplotrees are based on high probabilities.
On the tree picture is the Y SNP backbone of a tester, R > .. > R-L21 .. > R-FGC11134 > .. R-CTS4466 .. > R-A5313.
If a tester is positive for R-CTS4466, then he will most certainly be positive for R-FGC11134, for R-L21, and R-M269. If he falls neatly in a subclade of R-CTS4466, carrying subclade mutations, but he is NOT positive for, say, R-FGC11134, that could be because there was an imperfect copy of DNA from his father during transmission. This would be a rare event.
R-CTS4466 represents a subclade (child branch) of R-M269 so we say that R-CTS4466 is 👉 downstream from R-M269. R-M269 is a parent clade of R-CTS4466 so we say that R-M269 is 👉 upstream from R-CTS4466. Upstream SNPs are older and downstream SNPs are newer. On the leafy tree, the upstream SNPs are closer to the base of the tree, while the downstream SNPs are up in the thin branches.

FTDNA color codes its assumptions of your SNP mutations in its Y haplotree.
Green = tested positive.
Red = tested negative.
Blue = downstream.
Yellow = presumed positive.
Gray = presumed negative.
If you have not done any SNP testing, there will be no green or red SNPs in your haplotree. All the SNPs shown in the tree will be gray and yellow.
-
When you do a Y STR test, FTDNA comes up with a haplogroup prediction. The predictions for your matches are under the Y Haplogroup column. The predicted haplogroup is what appears if that tester has not done additional Y SNP testing.
When a match has done Y SNP testing, the Y Haplogroup is replaced by a new value - the terminal SNP. So the recorded SNP depends on how much Y SNP testing has been done.
The Y111 example shown to the right taken from the same data in CASE B1 under scenarios. Some table columns have been omitted.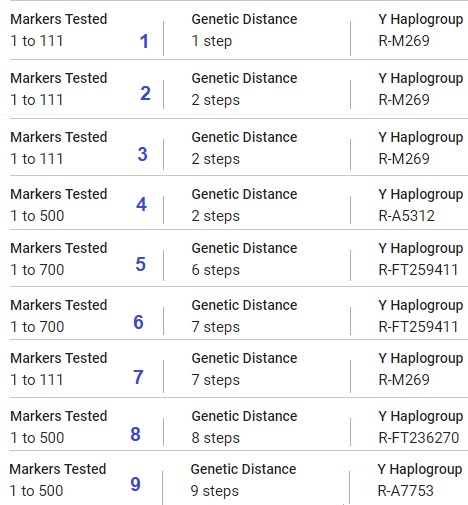
Y111 ExampleThe tester's predicted haplogroup is R-M269 (not shown in the table). He has not done further Y SNP testing. The same goes for Matches 1, 2, and 3; Match 1 is a Genetic Distance of 1, and Match 2 and Match 3 are a Genetic Distance of 2 so they are quite close from a genealogical perspective. So is Match 4, who HAS done advanced Y SNP testing; his Y Haplogroup is no longer just predicted at R-M269 but shows the terminal SNP R-A5312. Match 4 is in fact serving as an SNP testing proxy for his cousins. R-A5312 is downstream from the more well-known parent haplogroup R-CTS4466, which is downstream from R-M269.
The tester, plus Matches 1 through 4, are all descended from same-surnamed men from the same location. The necessary genealogy records for determining the most recent common ancestor don't exist and they don't know exactly the degrees of their relationships to each other. Autosomal DNA has not been helpful up this point, primarily because these men all have ancestry from the same close-knit rural area and are almost certainly related to each other in more than one way. One of these men even has the same surname-geographic area ancestry in his direct maternal line.
The relationship between these SNPs is R-M269 > R-CTS4466 > R-A5312. R-CTS4466 is downstream from R-M269 and upstream from R-A5312. R-A5312 is downstream from the all the others. Match 4 cannot do more advanced SNP testing to further explore this SNP branch, so we are unsure what an updated terminal SNP would be.
Matches 5 through 9 are a Genetic Distance of 6 or more steps away and are of different surnames. In this particular case, they can be ignored from a genealogical perspective.
Match 5 and Match 6 have done advanced Y SNP testing and show a terminal SNP R-FT259411. The SNP stream is: R-M269 > R-CTS4466 > R-A5312 > R-BY34895 > R-FT259411.
Match 8, at a Genetic Distance of 8, has a terminal SNP of R-FT236270. The SNP stream is: R-M269 > R-CTS4466 > R-A5312 > R-FT236270.
Match 9, at a Genetic Distance of 9, has a terminal SNP of R-A7753. The SNP stream is: R-M269 > R-CTS4466 > R-A5312 > R-BY23645 > R-A7753.
The Y67 example to the right is an expansion of CASE B2 under scenarios. Some columns have been omitted. Names and some terminal SNPs have been changed.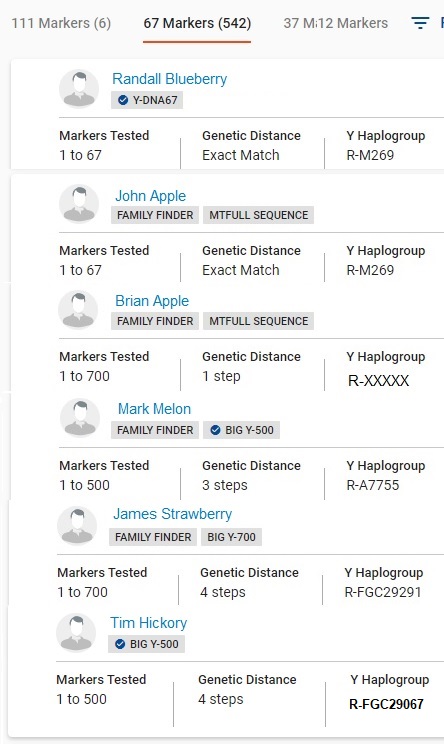
Y67 ExampleTester APPLE has well over500 matches at this level. Through genealogical research he knows he is second cousins with Brian APPLE (GD=1).
Randal Blueberry is believed to have experienced an NPE event in his lineage from the Apples (GD=0), but he has never responded to any inquiries. A search was made on Ancestry.com and his last known post was in 2010. Tester Apple and Brian Apple are third cousins to John Apple (GD=0), pushing the most recent known common Apple ancestor further back (born about 1783). Since Randal Blueberry's documented earliest known ancestor was born about 1818, and one of the Apples was much later known to have settled in close proximity to the Blueberrys, it looks more like the NPE direction was from Apple to Blueberry.
Tester Apple has also done Big Y testing and has Brian Apple's terminal SNP: R-XXXXX. No other related Apples have materialized through Y DNA testing. They exist in a subclade of R-CTS4466 > S1115.
Mark Melon (GD=3), James Strawberry (GD=4), and Tim Hickory (GD=4) all lie downstream of R-CTS4466 > S1115, conflicting with each other and with the subclade of the Apples:S1115 > FGC84010 > A541 > A1135 > FT7592 > A195 > A761 > A88 > Z16259 > A2220 > A7755
(Melon: GD = 3)
S1115 > FGC84010 > A541 > S1121 > Z16252 > Z18170 > FGC29280 > FGC29291
(Strawberry: GD=4)
S1115 > FGC84010 > A541 > S1121 > Z16252 > A9005 > FGC29068 > A804 > FGC29067
(Hickory: GD=4)Had the extra testing not been done on other Apple relatives and the advanced SNP testing done, Tester Apple might have assumed he was an NPE from one of these other surnames or surnames of matches that are not shown.
John Apple and Randal Blueberry have NOT done any advanced Y SNP testing. Therefore their predicted haplogroup R-M269 remains shown. Tester Apple and everyone else in the table have done advanced Y testing.
This is a superb example making a case for upgrading to Y111, getting known distant relatives tested, and for doing advanced Y SNP testing. The extra testing helps filter out irrelevant matches. Tester Apple and Brian Apple are both deceased. Had one of their Apple relatives not gotten them and Cousin John Apple to test, this information would have been lost forever.
-
Such a match is encouraging! It carries a high probability you share a common ancestor within the past millennium. You and your match would both need to test more markers to ensure the match persists to relatively recent times and to better determine how recent your relationship is. Sometimes excellent matches at 37 markers disappear at higher levels of testing. Look at your genealogy paper work to see if you can determine your common ancestor.
-
Even ruling out unknown non-paternity events, YES! Two men may be perfect matches with a most recent common ancestor dating several hundred years ago. Y SNP testing would confirm the relationship and make sure you are not doppelgangers (next question).
Usually, we expect some marker change over that time period but the reverse can happen and we might see extraordinary marker stability.
-
YES. This situation differs from the prior question, in which two people are related. You and the other person could accidentally resemble each other on your STR markers but be unrelated genealogically. You've heard of doppelgangers (twin strangers), this is a similar idea.
Though it is possible, a true doppelganger STR match would be rare. Y SNP testing would confirm the absence of a relationship.
Suppose you walk into a church and see two red-haired children sitting together in the first pew next to some adults. It would be easy and natural to assume they are siblings from the same family, but you don't know that unless you ask.
The common aspect of these situations is that two people happen to share a physical resemblance, but aren't related.
-
The timeframe that STRs are reliable as a means of defining recent genealogical family lines can vary. Generally, 21-25 generations back might be a good point in time to expect some deterioration. It can happen sooner, and it can happen much much later.
When Y STR testing was very new, the first twelve markers were considered fairly reliable in identifying certain haplotypes such as Irish Type II. The formation of the Irish Type II haplotype is considerably older than the aforementioned guideline of 21-25 generations.
STRs are considerably more volatile than SNPs. There is a reason why the Y SNPs are considered the patriline backbone.
-
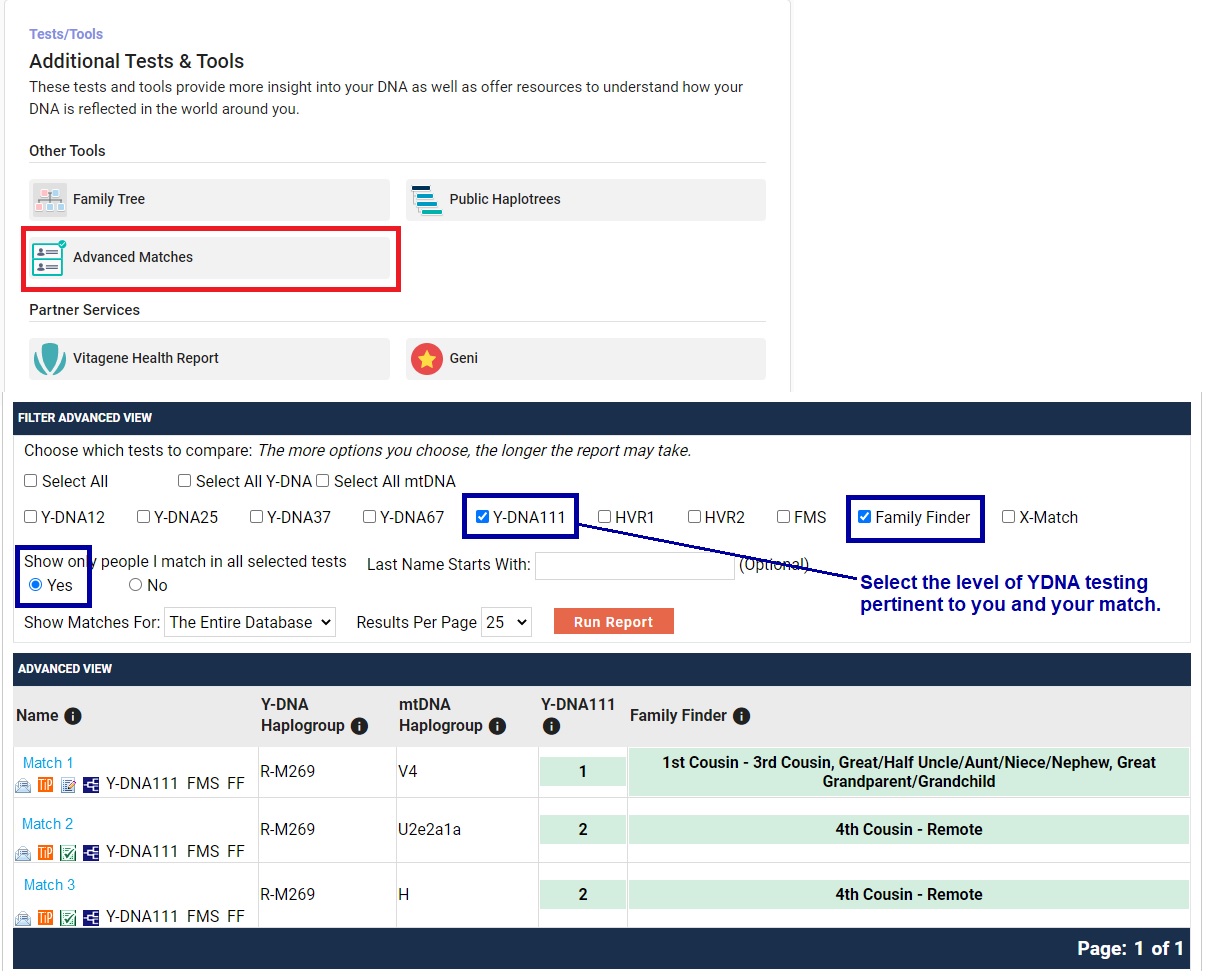
Y DNA Table OptionsAt the top of the Detail View of your Y DNA Matches table, you can select for the matches at a specific test level - 12, 37, 67, or 111 markers. There are also utilities for Filtering, Searching, and Sorting.


Y DNA Table Filtering OptionsFILTERING provides the following options: Genetic Distance; Groups (membership in your same projects); Family Tree documentation; Match Date; and Tests Taken.
Y DNA Table Search OptionsSEARCHING provides four options: by Name, by Y-DNA Haplogroup, by Paternal Country of Origin, or All.
Y DNA Table Sort OptionsSORTING provides many options: by Genetic Distatnce, by Markers Tested, by Big Y STR Differences, by Y-DNA Haplogroup, by Paternal Country of Origin, by Paternal Earliest Known Ancestor, and by Most Recent Matches.
-
A palindromic marker occurs on a Y chromosome location in which recombination events can occur within the Y chromosome. One part of the Y chromosome can write itself on top of another part. We recognize these markers as multi-copy markers.
There are a number of palindrome regions on the Y chromosome, labeled, P1, P2, P3, P4, P5, P8. Palindromic sequences of DNA form hairpin shapes.
Sometmes the two arms of the hairpin may eventually develop mismatches, messing up the base pairs. The cell uses enzymes to try repairing the mismatches. This could result in a successful repair, or it could result in yet another copy being made, or a copy being deleted.
DYS459, CDY, and DYS464 occur on P1. They normally contain 2, 2, and 4 markers respectively. If you see extra values on one of these markers beyond the number of markers we would ordinarily expect, there is a chance you'll see extra values on the other two markers. Repair attempts occurred on the hairpin, not just on one marker.
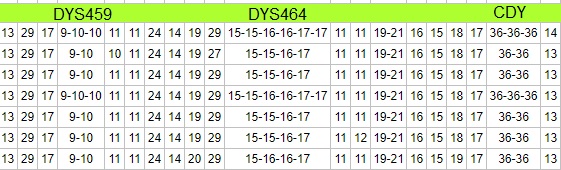
Extra values on these multi-copy markers can make two individuals look more remotely related when in fact they may not be so far apart. Their occurrence can wreak havoc on Genetic Distance calculations. In the example shown above, one could argue that the spawning of the extra values occurred as a consequence of a single hairpin repair event, rather than three separate mutations.
It is also possible for these multi-copy markers to delete copies.
For more details, see:
RecLOH (Recombinational Loss of Heterozygosity) by ISOGG.
Matching Multicopy Y-STR Markers In Closely Related Individuals by Thomas Krahn.
-
The short answer is that matches might appear at 67 and 111 markers that did not appear at 37 markers.
There may be more volatility in the markers, say, between 25 and 37 (Panel 3), and more stability after 37. Two markers in this panel called CDY and DYS576 are known to be volatile. FTDNA only displays matches for GD = 4 or less for 37 markers. If you are matching people at GD = 5 for 37 they would not be in your match list. But there could be exceptional stability in the markers from 38 to 67 so that matches who didn't show up at 37 might show up at 67 markers.
The illustration below shows the names of the first 37 markers, color coded for their relative volatility. More than 4 mutations that separate you apart from others, frequently occurring on those more volatile markers, will be enough to push you off each other's match lists.
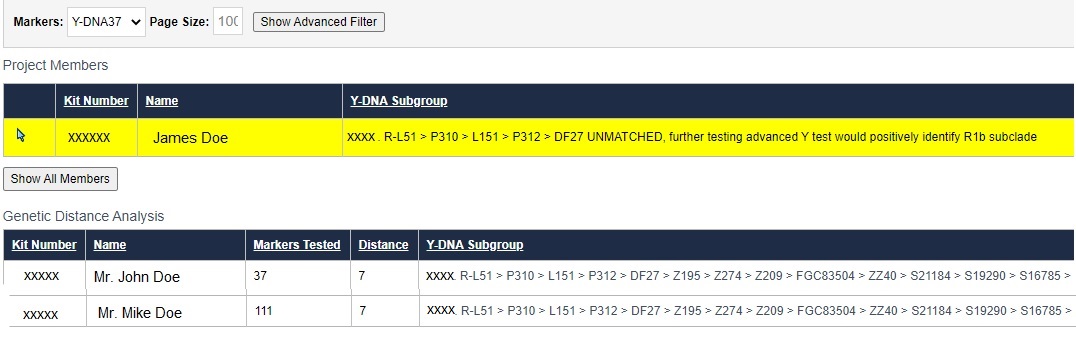
Here is an example with James Doe, a tester who has completed Y37 and has no relevant surname matches. The administrator has run a deeper Genetic Distance analysis. Prior to running the analysis the administrator already estimated that James falls under haplogroup R-DF27. The deeper analysis in this surname project reveals that, at 37 markers, he is GD=7 from John Doe, another R-DF27 project member who has tested Y37. In addition, there is a Mike Doe, who matches John Doe. Mike has tested the full Y111. Mike and John are grouped together as a lineage.
While there are no guarantees that James will match Mike at a higher number of markers, he won't know unless he upgrades his Y37 test. Recall also an earlier question that talked about FTDNA's match cutoff points. GD can be reexamined at 111 markers to get a better sense of how closely they are all related.
-
There could be an error with your results, or perhaps the Y chromosome was somehow damaged in your DNA. If you get such a result, ask FTDNA to check it again.
This is frustrating when this occurs, because you won't be able to match your known Y line cousins through your test result. If FTDNA says there is no error, you could consider submitting a different swab via a different account and trying the test again. It's up to you whether you want to incur the expense.
This is extremely rare.
-
Not normally. Should circumstances arise that the lab needs more of your sample, it sends you another test kit.
If you are supervising the Y test of a man who is unable to provide more swab sample, or is deceased, there is some risk in ordering an upgrade, in case that original submitted swab sample has deteriorated.
👉 Part IV explains why Big Y upgrades probably should NOT be ordered for testers who are not able to or are unwilling to submit another cheek swab.
-
Yes. For different parent haplogroups, e.g. I vs R, it is improbable they are related. For different subgroups deep in a parent haplogroup's descendant tree it is possible, e.g. R-A224 a branch of R-A223 deep in R. The yDNA matches can only say what is probable and you will have to decide from that was is possible. It is not unlike the decisions jurors have to make when contemplating reasonable doubt. Some numbers to consider:
Genetic Distance For 67 Markers Probability of a common ancestor within 3 generations 13 2 in a billion 10 0.00003% 5 0.3% 1 40% The probability is never zero. Hence no matter the genetic distance (GD) between two people in different haplogroups it is theoretically possible they are related. At GD=1 it is clearly possible, at GD=5 it rarely happens and at GD=10 it probably never happens for practical purposes. So where do you want to draw the line?
-
A parent haplogroup conflict between two lines of descent in a well documented old pedigree could indicate a non-paternity event occurring somewhere in one of the lines of descent. When a common ancestor lived say 300 or 400 years ago, then the NPE could have occurred many generations ago.
This is a difficult-to-impossible problem to resolve, especially if the NPE occurred too many generations ago so as to be beyond the practical reach of autosomal DNA matching.
To resolve such a conflict with Y DNA requires identifying living men who according to available documentation are in different patrilines of descent from the common ancestor, then doing Y DNA sampling on those men today in those patrilines. This assumes that those men exist, you are able to find them, and they are willing and able to test.
-
The FTDNA Learning Center gives more information on these topics.